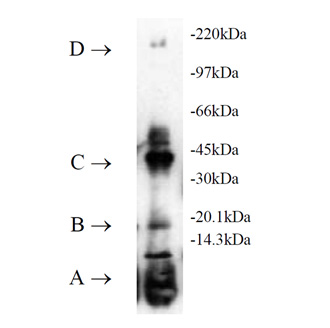
Ubiquitinylation of proteins constitutes an important cellular mechanism for targeting short-lived proteins for degradation by the 26S proteasome. Three classes of enzymes are involved in the conjugation of ubiquitin to proteins. E1, the ubiquitin activating enzyme, activates ubiquitin through the ATP-dependent formation of a high-energy thiol ester bond between the carboxyl terminus of ubiquitin and the active-site cysteine within E1. This E1-activated ubiquitin is transferred to a cysteine residue of an E2, or ubiquitin-conjugating enzyme (UbC). E2 enzymes, either by themselves or in conjunction with E3 enzymes (ubiquitin ligases), then transfer ubiquitin to target proteins forming stable isopeptide bonds resulting in multi-ubiquitin chain formation. It is the diverse combinations of E2-E3 complexes which are thought to define substrate specificity.
UbcH2 is the human homolog to the yeast DNA repair gene RAD6, which is induced by DNA damaging reagents. It has been shown to conjugate ubiquitin to histone H2A in an E3 dependent manner in vitro, work which clearly demonstrated that UbcH2 consists of two functionally independent domains, an N-terminal core domain with ubiquitin conjugating activity, and a C-terminal domain which interacts with substrate proteins. UbcH2 has been associated with cancer-induced cachexia and with the regulation of sepsis-induced muscle proteolysis.
Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
| Regulatory Status |
RUO – Research Use Only |
|---|
Last modified: May 29, 2024
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?