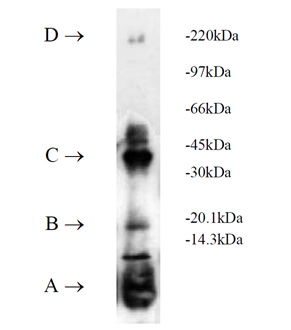
Ubiquitinylation of proteins constitutes an important cellular mechanism for targeting short-lived proteins for degradation by the 26S proteasome. Three classes of enzymes are involved in the conjugation of ubiquitin to proteins.
E1, the ubiquitin activating enzyme, activates ubiquitin through the ATP-dependent formation of a high-energy thiol ester bond between the carboxyl terminus of ubiquitin and the active-site cysteine within E1. This E1-activated ubiquitin is transferred to a cysteine residue of an E2, or ubiquitin-conjugating enzyme (UbC).
E2 enzymes, either by themselves or in conjunction with E3 enzymes (ubiquitin ligases), then transfer ubiquitin to target proteins forming stable isopeptide bonds resulting in multi-ubiquitin chain formation. It is the diverse combinations of E2-E3 complexes which are thought to define substrate specificity.
Among ubiquitin-conjugating enzymes, the mammalian ubiquitin conjugating enzyme UbcH1, also known as E2-25k, is unique in its ability to catalyse the synthesis in vitro of unanchored Lys48-linked poly-ubiquitin chains from mono- or poly-ubiquitin, E1, and ATP. The properties of truncated and chimeric forms of E2-25k suggest that the polyubiquitin chain synthesis activity of this E2 depends on specific interactions between its conserved 150-residue core domain and its unique 50-residue tail domain. Similarly E2-25k can catalyse the cyclisation of longer poly-ubiquitin chains, including tetra- and penta-ubiquitin. Although cyclic tri-ubiquitin resists hydrolysis by the PA700 or isopeptidase T deubiquitinylating enzymes, it can be disassembled to ubiquitin monomers by isopeptidase(s) in a red blood cell extract. Thus, if cyclic poly-ubiquitin forms in vivo, it will not accumulate as a dead-end product.
Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
| Regulatory Status |
RUO – Research Use Only |
|---|
Last modified: May 29, 2024
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?