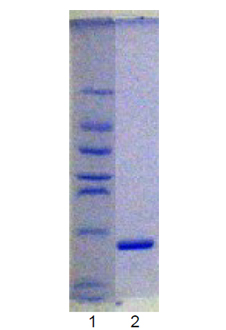
Ubc9, the only SUMO E2 enzyme, conjugates activated SUMO and mediates its subsequent linkage, via C-terminal isopeptide bond formation, to a number of proteins, including RanGAP1, SP100, p53, IκBα and PML, without the absolute requirement for an E3 ubiquitin-protein ligase-like activity. SUMOylation is involved in a range of processes including nuclear protein targeting, formation of sub-nuclear complexes, regulation of transcriptional activities, and control of protein stability.
The mechanism for SUMO conjugation is analogous to that of the ubiquitin system. Unlike ubiquitinylation, which leads inter alia to a degradative pathway, SUMO modification of target proteins is involved in nuclear protein targeting, formation of sub-nuclear complexes, regulation of transcriptional activities, and control of protein stability. A short sequence containing the consensus Ψ-K-X-D/E (where lysine is the modified amino acid, Ψ is a large hydrophobic residue and X is any amino acid residue) is thought to be necessary for protein SUMOylation to occur however SUMOylation has also been observed in cases where the consensus site is not conserved. Ubc9, the only SUMO E2 enzyme, conjugates activated SUMO (but not ubiquitin) and mediates its subsequent linkage, via C-terminal isopeptide bond formation, to a number of proteins, including RanGAP1, SP100, p53, IκBα and PML, without the absolute requirement for an E3 ubiquitin-protein ligase-like activity, at least in vitro. Direct interaction of UbcH9 with protein substrates, an unusual feature for an E2 conjugating enzyme, is thought to play a role in substrate recognition.The SUMO E1 activating enzyme heterodimer (Aos1/Uba2) is also required for initiation of the SUMOylation cascade.
Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
| Regulatory Status |
RUO – Research Use Only |
|---|
Last modified: May 29, 2024
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?