The proteasome is widely recognised as the central enzyme of non-lysosomal protein degradation. It is responsible for intracellular protein turnover and it is also critically involved in many regulatory processes and, in higher eukaryotes, in antigen processing. The 26S proteasome is the key enzyme of the ubiquitin/ATPdependent pathway of protein degradation. The catalytic core of this unusually large (2000kDa, 450Å in length) complex is formed by the 20S proteasome, a barrel shaped structure shown by electron microscopy to comprise of four rings each containing seven subunits. Based on sequence similarity, all fourteen 20S proteasomal subunit sequences may be classified into two groups, α and β, each group having distinct structural and functional roles. The α-subunits comprise the outer rings and the β-subunits the inner rings of the 20S proteasome. Observations of the eukaryotic proteasome and analysis of subunit sequences indicate that each ring contains seven different subunits (α7β7β7α7) with a member of each sub-family represented in each particle. Each subunit is located in a unique position within the α- or β-rings. In addition to the 20S particle, the 26S complex contains over twenty additional proteins, ranging in molecular weight from 25 to 10kDa, located in a distinct complex called the ‘PA700 proteasome activator’ or the ‘19S complex’, and which determines substrate specificity and provides the multiple enzymatic functions necessary for proteolysis and viability. Systematic analysis of the sub-unit components have revealed at least six members to be ATPases belonging to a new family of ATP-binding proteins, together with a further fifteen subunits that lack the capacity to bind ATP, isopeptidases and several other proteins thought to be responsible for the unfolding of a protein substrate prior to insertion into the proteolytic core of the 20S proteasome.
Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
This antibody is covered by our Worry-Free Guarantee.
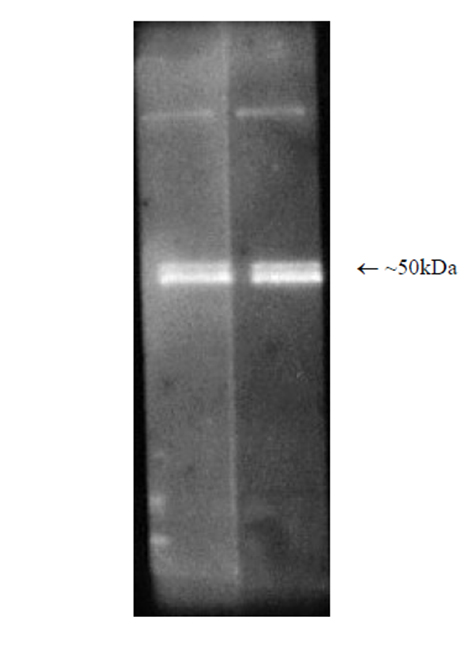
Western blot analysis: Luminograph of rabbit 26S proteasome preparation after SDS PAGE followed by blotting onto PVDF membrane and probing with antibody BML-PW8770. Antibody dilution 1:1000 and 1:2500 using ECL procedure (1 min exposure).
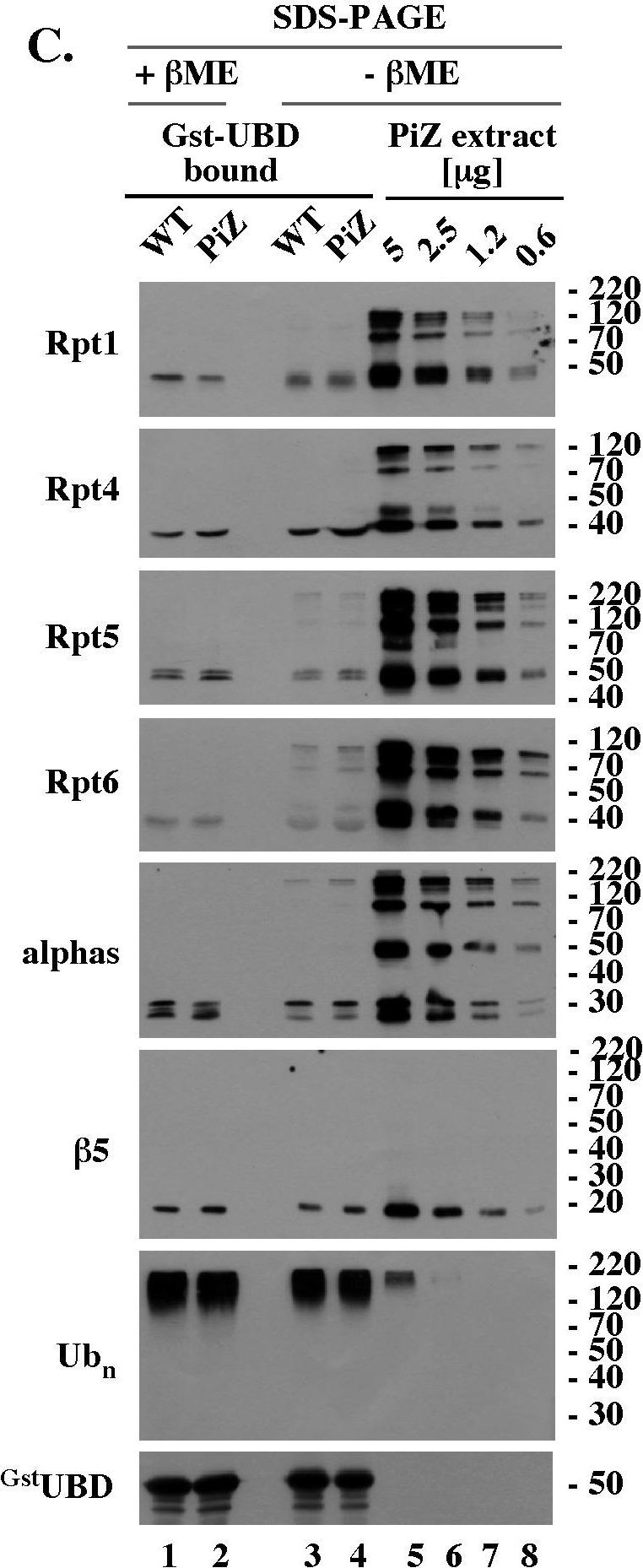
Reduction-sensitive modifications typical of aging WT mice accumulate prematurely on selected proteasomal subunits in the livers of PiZ mice.(A). Rpt4 Western blot analysis of unreduced, un-boiled liver extracts. 5 µg of the indicated extracts were mixed with Laemmli buffer without (unreduced samples) or with (reduced samples) βME (−/+ βME), separated by SDS-PAGE without prior boiling, and analyzed by Western blot with antibodies specific to Rpt4. (B). Rpt4 Western blot analysis of unreduced, but boiled, samples. Experiment like in A, lanes 1–9, except that extracts were mixed with Laemmli buffer without βME (- βME) and boiled for 4 minutes prior to SDS-PAGE. (C). Analysis of 26S proteasomes co-purified with polyubiquitin conjugates. WT and PiZ 26S proteasomes were co-purified with polyubiquitin conjugates as described in Fig. 4B and analyzed by Western blot after separation by SDS-PAGE with (lanes 1, 2) and without (lanes 3, 4) prior reduction by βME. Serial dilutions of unreduced liver extract from 103 old PiZ mouse are shown as reference (lanes 5–8). Extract prepared from 103 days old WT mice had similar reduction-sensitive modifications (data not shown, see panel A). (D). HPLC of WT and PiZ liver extracts followed by unreduced SDS-PAGE/Western blot analysis. Experiment like Fig. 5B, except that gel filtration fractions were not reduced with βME before SDS-PAGE. Data shown in A-D are representative of at least 3 independent experiments.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: PiZ mouse liver accumulates polyubiquitin conjugates that associate with catalytically active 26S proteasomes. PLoS One (2014)
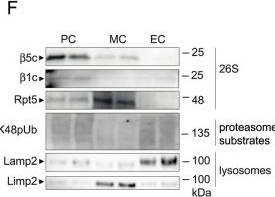
Glomerular cell-specific distribution of proteasomal and lysosomal proteins in human and murine glomeruli.A Schematic depiction of the ubiquitin proteasome system (UPS) and autophagosome-lysosome pathway (ALP) with individual marker proteins used for the expression analyses within glomerular cell types. Ubiquitin = polypeptide involved in dynamic regulation of protein function, localization, and stability; Rpt5 = proteasome regulatory subunit 6A (Psmc3) of the proteasome 19S regulatory particle; β5c (Psmb5) = main proteolytic subunit of the constitutive 20S core particle; LC3 = microtubule-associated protein 1A/1B-light chain 3; Lamp1 and Lamp2 = lysosomal-associated membrane proteins 1 or 2; Limp2 (Scarb2) = lysosomal integral membrane protein 2. Distribution of marker proteins (green) by high-resolution confocal images in a healthy human (B) and murine (C) glomerulus in relation to the slit diaphragm protein nephrin (red) and DNA (blue); pc podocyte, mc mesangial cell, ec glomerular endothelial cell, pec parietal epithelial cell, white arrows point toward endothelial lining filled with Lamp2-positive lysosomes, n = 3 individuals. D–F Podocytes (PC), mesangial cells (MC) and glomerular endothelial cells (EC) were bulk-isolated from glomeruli. D Proteomic analyses depict molecular properties of glomerular cell types as shown by the radar plot, whereby two-fold changes of distinct uniprot key words are plotted. Protein values were obtained by label-free quantification results using the MaxQuantLFQ algorithm72. E Relative transcript levels quantified via qRT-PCR of Psmb5 (encoding for β5c) and Scarb2 (encoding for Limp2) normalized to 18S as home keeper in relation to total glomerular transcript levels (dashed line), mean ± SEM, *p = 0.0292 (PC Psmb5), *p = 0.048 (MC Scarb2), one-way ANOVA with Bonferroni post-test for multiple comparisons, n = 12 of 2 pooled independent experiments. F Protein abundance from isolated glomerular cell types determined by immunoblot, equal loading was ensured by loading equal numbers of FACS-sorted PCs, MCs, and ECs, n = 3 independent experiments. Scheme was created with BioRender.com. Source data are provided as a Source Data file.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: The proteasome modulates endocytosis specifically in glomerular cells to promote kidney filtration. Nat Commun (2024)
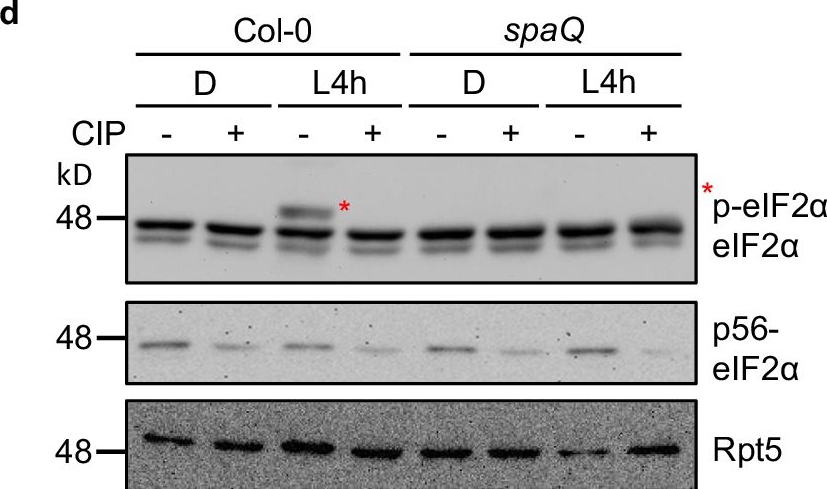
SPA1 phosphorylates C-terminus of eIF2α.1 and eIF2α.2 in a light-dependent manner.a Predicted phosphorylation sites located at C-terminal eIF2α are indicated in red. Tyr, Ser or Thr were then replaced with alanine (A) and aspartic acid (D) to generate a phospho-null form and a phospho-mimicking form of eIF2α, respectively. b A semi-in vivo kinase assay showed that SPA1 protein extracted from TAP-SPA1 overexpression line phosphorylates wild-type (WT) eIF2α but not the phospho-null form (6A/8A) of eIF2α (autoradiogram in upper panel). The plant extracts were derived from 4-day-old TAP-SPA1 seedlings incubated either under dark (D) or illuminated under white light for 4 h (L4h) before sampling. Three biological repeats of data showed the same results. c A semi-in vivo kinase assay showed that WT version of eIF2α has higher phosphorylation intensity in TAP-SPA1 than in spaQ under light condition (autoradiogram in upper panel). The plant extracts were derived from 4-day-old TAP-SPA1 and spaQ incubated either under dark (D) or illuminated with 4 h white light (L4h) before sampling. The GST only was used as negative control. The lower panel shows the protein levels in a Coomassie blue staining gel. Three biological repeats of data showed the same results. d Immunoblots showed phosphorylation of eIF2α under both dark and light condition in Col-0 and spaQ. Four-day-old Col-0 and spaQ etiolated seedlings incubated either under dark (D) or illuminated with 4 h white light (L4h) before sampling, and extracted proteins were then separated on 6.5% sodium dodecyl sulfatepolyacrylamide gel electrophoresis (SDS-PAGE) gels containing 15 μM Phos-tag. The slow-migrating band (*) is the phosphorylated form of eIF2α. Three biological repeats of data showed the same results. Treatment of calf intestinal alkaline phosphatase (CIP, +) and inactivated boiled CIP (-) were shown. Rpt5 proteins were used as loading control.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: The phosphorylation of carboxyl-terminal eIF2α by SPA kinases contributes to enhanced translation efficiency during photomorphogenesis. Nat Commun (2024)




Product Details
| Alternative Name |
TBP1, Tat-binding protein 1, 26S protease regulatory subunit 6A, Proteasome 26 S subunit ATPase 3 |
|---|---|
| Application |
IHC, WB |
| Clone |
TBP1-19 |
| Formulation |
Liquid. Contains 10mM sodium azide. |
| Host |
Mouse |
| Immunogen |
Recombinant full-length human Rpt5. |
| Isotype |
IgG2b |
| Purity Detail |
Partially purified ascites. |
| Source |
Purified from hybridoma tissue culture supernatant. |
| Species Reactivity |
Human, Mouse, Rabbit, Rat |
| Specificity |
Recognizes the Rpt5/S6a subunit of the 19S regulatory subunit |
| Technical Info / Product Notes |
The hybridoma secreting the antibody to subunit Rpt5/S6a was generated by fusion of splenocytes from Balb/c mice that had received repeated immunisation with a full length human recombinant protein with Sp2/0-Ag14 myeloma cells. The antibody (clone TBP1-19) has been characterised by one-dimensional Western blotting. Immunoblotting – Single dimension SDS-PAGE of a purified rabbit 26S proteasome preparation followed by Western blotting at optimal dilution (1:2500) gives a major band with a relative molecular weight of approximately 50kDa. There is a minor band observed running slightly slower that may be due to modification. |
| UniProt ID |
P17980 |
| Worry-free Guarantee |
This antibody is covered by our Worry-Free Guarantee. |
Handling & Storage
| Long Term Storage |
-20°C |
|---|---|
| Shipping |
Blue Ice |
| Regulatory Status |
RUO – Research Use Only |
|---|
- Deciphering the role of autophagy under Cd toxicity in Arabidopsis thaliana: Collado-Arenal, A. M., Pérez-Gordillo, F. L., et al.; bioRxiv , (2025), Application(s): Western Blot
- Cypin regulates K63-linked polyubiquitination to shape synaptic content: Gandu, S., Rodriguez, A. R., et al.; Sci. Adv. 11, eads5467 (2025), Abstract
- The proteasome maturation factor POMP moonlights as a stress-induced transcriptional regulator: Giandomenico, S. L., Mueller, M., et al.; bioRxiv , (2025), Application(s): Western blot
- The proteasome modulates endocytosis specifically in glomerular cells to promote kidney filtration: Sachs, W., Heintz, L., et al.; Nat. Commun. 15, 1897 (2024), Application(s): WB, Abstract
- The phosphorylation of carboxyl-terminal eIF2α by SPA kinases contributes to enhanced translation efficiency during photomorphogenesis: Chang, H. H., Huang, L. C., et al.; Nat. Commun. 15, 3467 (2024), Application(s): WB, Abstract
- COP1 controls light-dependent chromatin remodeling: Wang, W., Kim, J., et al.; PNAS 121, e2312853121 (2024), Application(s): WB, Abstract
- The Proteasome Modulates Endocytosis in a Glomerular Cell Type Specific Modality to Secure Kidney Filtration: Sachs, W., Blume, L., et al.; Research Square , (2023)
- The Arabidopsis Deubiquitylase OTU5 Suppresses Flowering by Histone Modification-Mediated Activation of the Major Flowering Repressors FLC, MAF4, and MAF5: R. Radjacommare, et al.; Int. J. Mol. Sci. 24, 6176 (2023), Abstract
- PP2A Dephosphorylates Phytochrome-Interacting Factor 3 to Modulate Photomorphogenesis inArabidopsis: Cai, X., Lee, S., et al.; bioRxiv , (2023)
- Raising cGMP restores proteasome function and myelination in mice with a proteotoxic neuropathy.: Goldberg, A. L., Wrabetz, L., et al.; Brain 145, 168 (2022), Application(s): WB, Abstract
- A versatile new tool derived from a bacterial deubiquitylase to detect and purify ubiquitylated substrates and their interacting proteins: M. Zhang, et al.; PLoS Biol. 20, e3001501 (2022), Abstract
- Direct phosphorylation of HY5 by SPA kinases to regulate photomorphogenesis in Arabidopsis.: Wang, W., Paik, I., et al.; New Phytol. 230, 2311 (2021), Application(s): WB, Abstract
- Spatial regulation of thermomorphogenesis by HY5 and PIF4 in Arabidopsis: S. Lee, et al.; Nat. Commun. 12, 3656 (2021), Application(s): WB, Abstract
- Phytochrome B triggers light-dependent chromatin remodelling through the PRC2-associated PHD finger protein VIL1: J. Kim, et al.; Nat. Plants 7, 1213 (2021), Application(s): WB, Abstract
- Changes in the cellular fatty acid profile drive the proteasomal degradation of α-synuclein and enhance neuronal survival.: Xylaki, M., Boumpoureka, I., et al.; FASEB J. 34, 15123 (2020), Application(s): WB, Abstract
- Direct phosphorylation of HY5 by SPA1 kinase to regulate photomorphogenesis in Arabidopsis: Wang, W., Kim, J., et al.; bioRxiv , (2020), Application(s): WB
- Ecm29-mediated proteasomal distribution modulates excitatory GABA responses in the developing brain: Lee, M., Liu, Y. C., et al.; J. Cell Biol. 219, (2020), Abstract
- RhoJ integrates attractive and repulsive cues in directional migration of endothelial cells: Fukushima, Y., Nishiyama, K., et al.; EMBO J. 39, e102930 (2020), Abstract
- A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis.: Ngoc Pham, V., Huq, E., et al.; Nat. Commun. 10, 4216 (2019), Application(s): WB, Abstract
- Protein Expression of Proteasome Subunits in Elderly Patients with Schizophrenia: Scott, M. R., Rubio, M. D., et al.; Neuropsychopharmacology 41, 896 (2016), Abstract
- Skeletal muscle myotubes in severe obesity exhibit altered ubiquitin-proteasome and autophagic/lysosomal proteolytic flux: L.M. Bollinger, et al.; Obesity (Silver Spring) 23, 1185 (2015), Application(s): Western Blot, Abstract — Full Text
- Bortezomib Amplifies Effect on Intracellular Proteasomes by Changing Proteasome Structure: D.S. Pitcher, et al.; EBioMedicine 2, 642 (2015), Application(s): Western Blot, Abstract
- Human ASPL/TUG interacts with p97 and complements the proteasome mislocalization of a yeast ubx4 mutant, but not the ER-associated degradation defect: Madsen, L., Molbæk, K., et al.; BMC Cell Biol. 15, 31 (2014), Abstract
- PiZ mouse liver accumulates polyubiquitin conjugates that associate with catalytically active 26S proteasomes.: Blomenkamp, K., Teckman, J., et al.; PLoS One 9, e106371 (2014), Application(s): WB, Abstract
- Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates.: Besche, H. C., Sha, Z., et al.; EMBO J. 33, 1159 (2014), Application(s): WB / Reactant(s): Human, Abstract
- Nuclear proteasomes carry a constitutive posttranslational modification which derails SDS-PAGE (but not CTAB-PAGE): D.S. Pitcher, et al.; Biochim. Biophys. Acta 1844, 2222 (2014), Abstract
- Reduction in ATP levels triggers immunoproteasome activation by the 11S (PA28) regulator during early antiviral response mediated by IFNβ in mouse pancreatic β-cells.: Buller, R. M., Corbett, J. A., et al.; PLoS One 8, e52408 (2013), Application(s): WB, Abstract
- TRIM5α associates with proteasomal subunits in cells while in complex with HIV-1 virions: Z. Lukic, et al.; Retrovirology 8, 93 (2011), Abstract — Full Text
- BAG-6 is essential for selective elimination of defective proteasomal substrates: Minami, R., Hayakawa, A., et al.; J. Cell Biol. 190, 637 (2010), Abstract
Last modified: May 29, 2024
Datasheet, Manuals, SDS & CofA
Manuals And Inserts
Certificate of Analysis
Please enter the lot number as featured on the product label
SDS
Enzo Life Science provides GHS Compliant SDS
If your language is not available please fill out the SDS request form
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?
