The 104 kDa yeast heat shock protein (Hsp104) is a cytosolic member of the Hsp100 family of proteins. Hsp104 cooperates with Hsp40 and Hsp70 co-chaperones in yeast in the reactivation of heat-damaged proteins. Hsp104 is also critical for the establishment and maintenance of the [PSI] prion phenotype in Saccharomyces cerevesiae.
Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
This antibody is covered by our Worry-Free Guarantee.
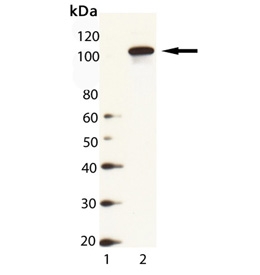
Western Blot Analysis of HSP104 (yeast), pAb (Prod. No. ADI-SPA-1040): Lane 1: MW Marker, Lane 2: Yeast Cell Lysate
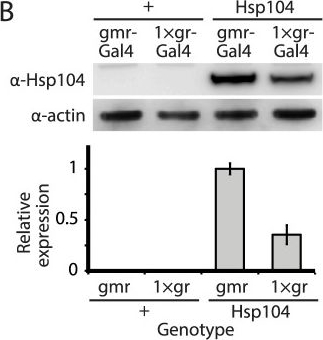
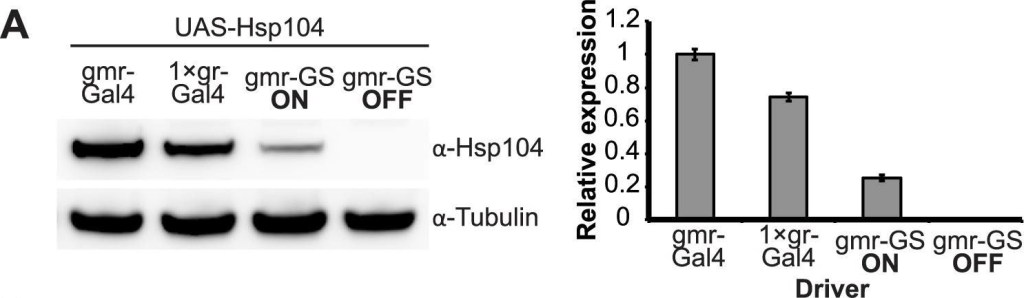
Tuning Hsp104 expression level for the fly eye.(A) Expression of UAS-Hsp104 using gmr-GAL4 caused disruption of internal retinal structure by d7. A less strong driver, 1×gr-GAL4 minimized this effect and prevented disruption to cellular organization within the retina. Arrows indicate the width of the retina to highlight changes in tissue integrity. (B) Immunoblots demonstrated that 1×gr-GAL4 drives lower Hsp104 expression than gmr-GAL4 at d7. Actin served as a loading control. Quantitation of immunoblots determined that 1×gr-GAL4 levels of Hsp104 were ∼35% that of gmr-GAL4. Hsp104 levels were normalized to actin (n = 3 (mean ± SEM)).
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)
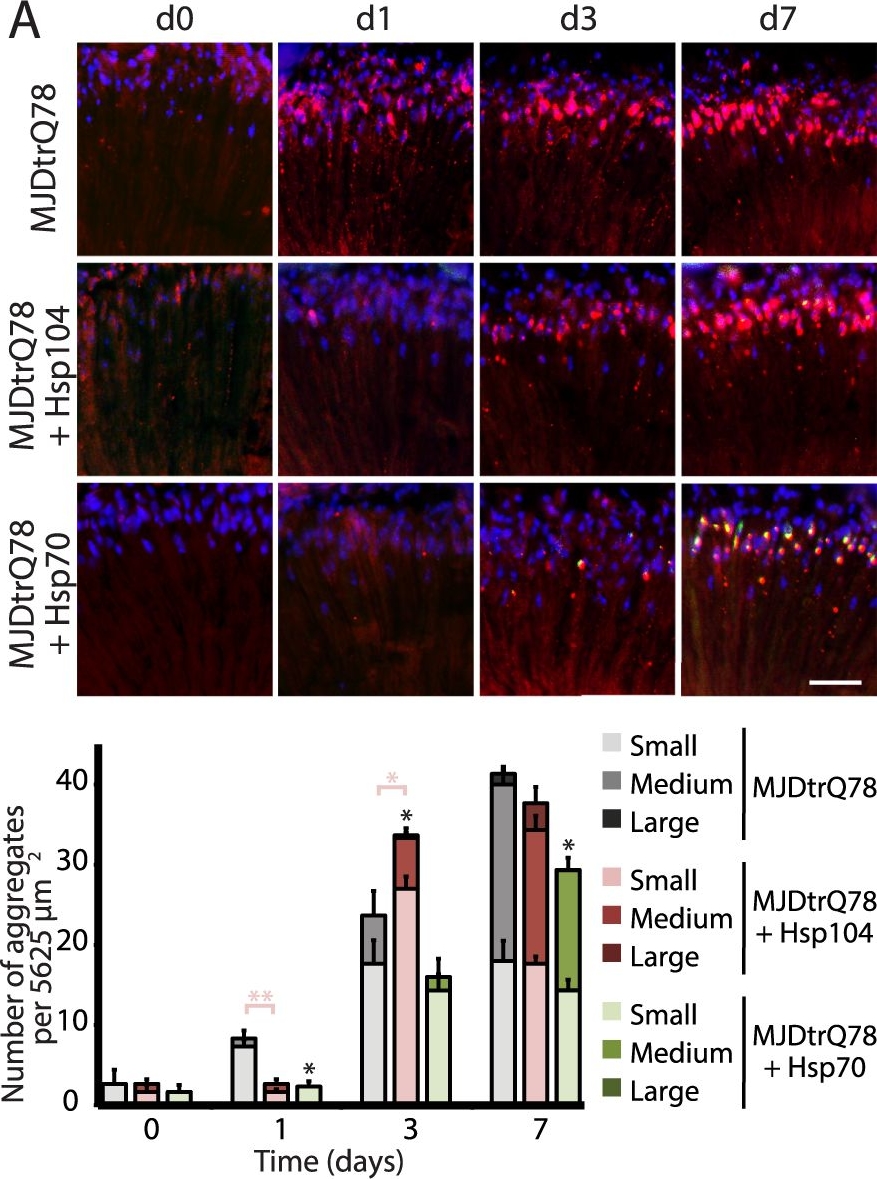
Hsp104 delays aggregation of truncated MJD, but enhances aggregation of full-length MJD.(A and B) With the rh1-GAL4 driver at indicated time points, cryosections and IHC demonstrate accumulations of MJDtrQ78 and MJDnQ78 (red) over time, using anti-HA and anti-myc antibodies, respectively. Sections were co-stained with anti-Hsp104 or anti-Hsp70 (green) as indicated, and nuclei were labeled by Hoechst (blue). Hsp104 delayed but did not suppress aggregation of MJDtrQ78, but Hsp104 enhanced accumulation formation of MJDnQ78. Hsp70 suppressed aggregation of both MJD proteins. The size of the aggregates was quantified using ImageJ, with delineations for large inclusions (>5 µm across), medium inclusions (2.5–5 µm), or small inclusions (<2.5 µm) (n = 3 (mean ± SEM)). Scale bar = 20 µm. *p<0.05, **p = 0.001–0.01, ***p<0.001; Statistics indicate comparison to the disease protein alone for total number of inclusions at each timepoint (black asterisks). Additional statistical comparisons for inclusion size divisions are indicated by color, e.g., dark red asterisk indicates significant change in large inclusions. (C and D) With the rh1-GAL4 driver at indicated time points, SDD-AGE and immunoblot analysis show the progression of amyloid formation of MJDtrQ78 and MJDnQ78 proteins over time. Hsp104 did not greatly affect the aggregation profile of MJDtrQ78 but enhanced formation of SDS-insoluble amyloid aggregates of MJDnQ78. Hsp70 suppressed aggregation of both MJD proteins. The formation of large, SDS-insoluble aggregates by SDD-AGE corresponded with the disappearance of SDS-soluble soluble protein from immunoblots. MJDtrQ78 and MJDnQ78 were detected using anti-HA and anti-myc, respectively, with anti-tubulin as a loading control. Band density for both amyloid smears (SDD-AGE) and soluble bands (Western blot) were quantified using ImageJ (n = 3 (mean ± SEM)). *p<0.05, **p = 0.001–0.01, ***p<0.001; Statistics indicate comparison to the disease protein alone at each timepoint.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)
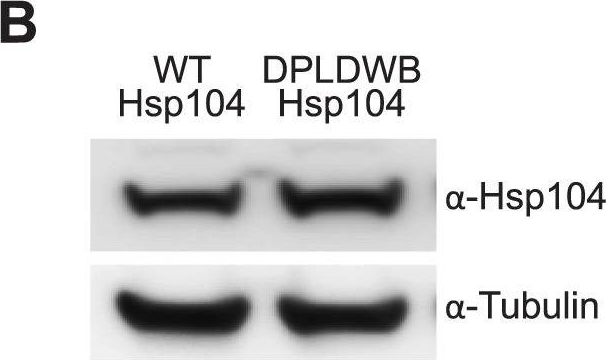
ATPase activity and substrate binding are required for Hsp104 to modulate disease.(A) With the gmr-GAL4 driver at d7, expression of the inactive mutant UAS-Hsp104DPLDWB, which is unable to bind substrate or hydrolyze ATP, caused no effect on its own when expressed by gmr-GAL4. Additionally, the inactive Hsp104DPLDWB did not modulate the toxicity of MJDtrQ78 or MJDnQ78. Eye images and retinal sections showed moderate degeneration upon expression of MJDtrQ78 or MJDnQ78, but unlike wild-type Hsp104 (see Fig. 2), Hsp104DPDLWB did not mitigate the degeneration caused by MJDtrQ78 nor did it enhance the toxicity of MJDnQ78. Arrows indicate the width of the retina to highlight changes in tissue integrity. (B) Western immunoblot demonstrated that WT Hsp104 and Hsp104DPLDWB had similar expression levels. Tubulin served as a loading control. Quantification of Western immunoblots confirmed that protein expression levels were similar, but that Hsp104DPLDWB was expressed at levels slightly higher than WT Hsp104. Hsp104 signal was normalized to tubulin (n = 3 (mean ± SEM)).
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)
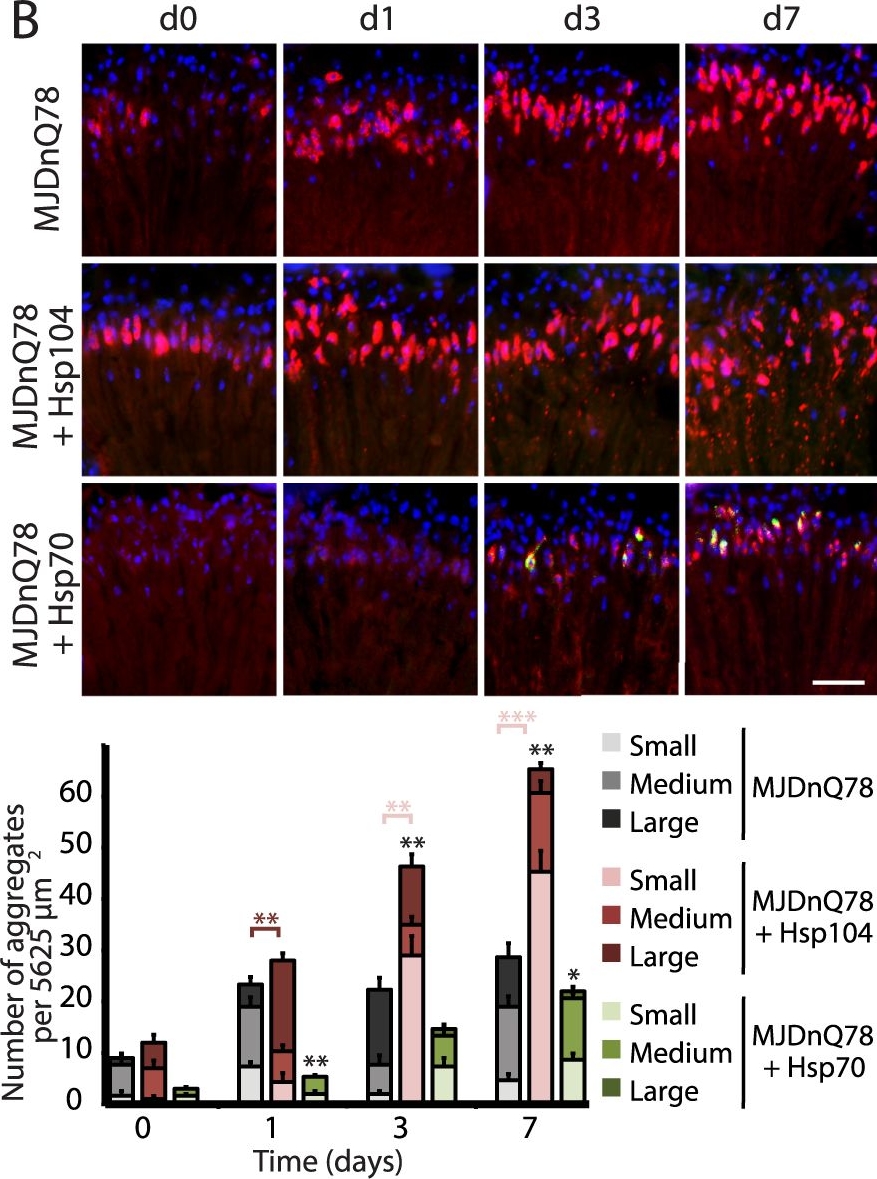
Hsp104 delays aggregation of truncated MJD, but enhances aggregation of full-length MJD.(A and B) With the rh1-GAL4 driver at indicated time points, cryosections and IHC demonstrate accumulations of MJDtrQ78 and MJDnQ78 (red) over time, using anti-HA and anti-myc antibodies, respectively. Sections were co-stained with anti-Hsp104 or anti-Hsp70 (green) as indicated, and nuclei were labeled by Hoechst (blue). Hsp104 delayed but did not suppress aggregation of MJDtrQ78, but Hsp104 enhanced accumulation formation of MJDnQ78. Hsp70 suppressed aggregation of both MJD proteins. The size of the aggregates was quantified using ImageJ, with delineations for large inclusions (>5 µm across), medium inclusions (2.5–5 µm), or small inclusions (<2.5 µm) (n = 3 (mean ± SEM)). Scale bar = 20 µm. *p<0.05, **p = 0.001–0.01, ***p<0.001; Statistics indicate comparison to the disease protein alone for total number of inclusions at each timepoint (black asterisks). Additional statistical comparisons for inclusion size divisions are indicated by color, e.g., dark red asterisk indicates significant change in large inclusions. (C and D) With the rh1-GAL4 driver at indicated time points, SDD-AGE and immunoblot analysis show the progression of amyloid formation of MJDtrQ78 and MJDnQ78 proteins over time. Hsp104 did not greatly affect the aggregation profile of MJDtrQ78 but enhanced formation of SDS-insoluble amyloid aggregates of MJDnQ78. Hsp70 suppressed aggregation of both MJD proteins. The formation of large, SDS-insoluble aggregates by SDD-AGE corresponded with the disappearance of SDS-soluble soluble protein from immunoblots. MJDtrQ78 and MJDnQ78 were detected using anti-HA and anti-myc, respectively, with anti-tubulin as a loading control. Band density for both amyloid smears (SDD-AGE) and soluble bands (Western blot) were quantified using ImageJ (n = 3 (mean ± SEM)). *p<0.05, **p = 0.001–0.01, ***p<0.001; Statistics indicate comparison to the disease protein alone at each timepoint.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)
Establishing the GeneSwitch paradigm.(A) Western immunoblot demonstrated that the gmr-GS-GAL4 line specifically drove Hsp104 expression in the presence of RU486 (gmr-GS ON), but there was no expression of Hsp104 in the absence of the drug (gmr-GS OFF) at d7. The level of expression by gmr-GS was lower than with the other eye-specific drivers gmr-GAL4 and 1×gr-GAL4. Tubulin served as a loading control. Quantification of Western immunoblots confirmed that gmr-GS expressed at a lower level than the other drivers, with the amount of Hsp104 expressed by gmr-GS reaching about 33% of that expressed by 1×gr-GAL4. Hsp104 levels were normalized to tubulin (n = 3 (mean ± SEM)). (B) Paraffin sections demonstrate that the retinal tissue loss associated with gmr-MJDtrQ78 is apparent at d0, and progresses through d7. In comparison, control flies (7 d) display no such loss of retinal integrity. For each example shown here, a 7,000 µm2 rectangular selection (used for quantification in Fig. 10) of a retinal section from three independent animals is presented. Each region was converted to a black and white image to show the area covered by tissue and quantitated by ImageJ analysis (see Methods). For the analysis in Fig. 10, regions from 6 independent animals were used for quantitation; all experiments were repeated at least three times with similar results.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)
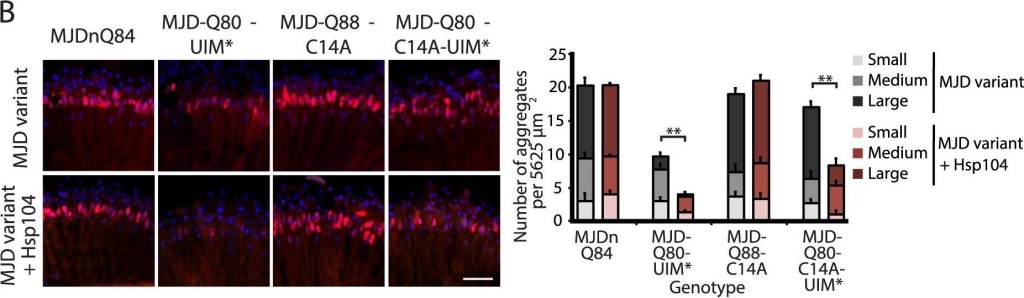
A portion of the Josephin domain and the Ubiquitin-Interacting Motifs prevent Hsp104 from rescuing full-length MJD pathogenicity.(A) With the 1×gr-GAL4 driver at d7, external eye and internal retinal structure showed suppression of toxicity by Hsp104 for MJD variants with mutated UIMs (MJD-Q80-UIM* and MJD-Q80-C14A-UIM*). Hsp104 strongly suppressed the external eye degeneration and loss of internal retinal structure of MJD lacking a region spanning the Josephin domain (amino acids 9–63 (Δ exon 2)). The MJDnQ84 and MJD-Q80-UIM* crosses were performed at 29°C to enhance the severity of degeneration. Arrows indicate the width of the retina to highlight changes in tissue integrity. (B) With the rh1-GAL4 driver at d3, Hsp104 suppressed inclusion formation in MJD variants with UIM mutations, as seen by IHC (d3). Accumulations of the MJD variant proteins were detected by anti-myc (red) and nuclei are labeled by Hoechst stain (blue). Scale bar = 20 µm. Size of inclusions was quantified using ImageJ, with delineations for large inclusions (>5 µm across), medium inclusions (2.5–5 µm), or small inclusions (<2.5 µm) (n = 3 (mean ± SEM)). Scale bar = 20 µm. **p = 0.001–0.01. (C) With the rh1-GAL4 driver at d3, SDD-AGE and Western immunoblot showed that by d3, Hsp104 enhanced aggregation of MJD variants with wild-type UIMs, but reduced formation of amyloid of MJD variants with UIM mutations. MJD variants were detected using anti-myc with anti-tubulin as a loading control. Band density for both amyloid smears (SDD-AGE) and soluble bands (Western blot) were quantified using ImageJ (n = 3 (mean ± SEM)). **p = 0.001–0.01.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model. PLoS Genet (2013)







Product Details
| Alternative Name |
ClpA, Heat shock protein 104 |
|---|---|
| Application |
WB |
| Application Notes |
Detects a band of ~104kDa by Western blot. |
| Formulation |
Liquid. In PBS containing 50% glycerol and 0.09% sodium azide. |
| GenBank ID |
Z73131 |
| Host |
Rabbit |
| Immunogen |
Synthetic peptide corresponding to the sequence near the C-terminus of yeast Hsp104. |
| Purity Detail |
Protein A affinity purified. |
| Recommendation Dilutions/Conditions |
Western Blot (1:1,000, ECL)Suggested dilutions/conditions may not be available for all applications.Optimal conditions must be determined individually for each application. |
| Source |
Purified from rabbit serum. |
| Species Reactivity |
Yeast |
| UniProt ID |
P31539 |
| Worry-free Guarantee |
This antibody is covered by our Worry-Free Guarantee. |
Handling & Storage
| Handling |
Avoid freeze/thaw cycles. |
|---|---|
| Long Term Storage |
-20°C |
| Shipping |
Blue Ice |
| Regulatory Status |
RUO – Research Use Only |
|---|
- Design principles to tailor Hsp104 therapeutics: Lin, J., Carman, P. J., et al.; Cell Rep. 43, 115005 (2024), Abstract
- Tuning Hsp104 specificity to selectively detoxify α-synuclein: Mack, K. L., Kim, H., et al.; Mol. Cell 83, 3314 (2023), Abstract
- Genetic inactivation of essentialHSF1reveals an isolated transcriptional stress response selectively induced by protein misfolding: Ciccarelli, M., Masser, A. E., et al.; bioRxiv , (2023)
- Genetic inactivation of essential HSF1 reveals an isolated transcriptional stress response selectively induced by protein misfolding.: Ciccarelli, M., Masser, A. E., et al.; Mol. Biol. Cell 34, ar101 (2023), Reactant(s): Saccharomyces cerevisiae, Abstract
- Dynamic coalescence of yeast Heat Shock Protein genes bypasses the requirement for actin.: Rubio, L. S., Gross, D. S., et al.; Genetics 223, (2023), Reactant(s): Saccharomyces cerevisiae, Abstract
- Molecular determinants and modifiers of Matrin-3 toxicity, condensate dynamics, and droplet morphology: M.L. Sprunger, et al.; iScience 25, 103900 (2022), Reactant(s) Saccharomyces cerevisiae, Abstract
- Rbs1 protein, involved in RNA polymerase III complex assembly in the yeast Saccharomyces cerevisiae, induces a Gcn4 response and forms aggregates when overproduced: I. Rudzińska, et al.; Gene 809, 146034 (2022), Abstract
- Wine Yeast Cells Acquire Resistance to Severe Ethanol Stress and Suppress Insoluble Protein Accumulation during Alcoholic Fermentation.: Yoshida, M., Furutani, N., et al.; Microbiol. Spectr. 10, e0090122 (2022), Application(s): WB / Reactant(s): Saccharomyces cerevisiae, Abstract
- Acquired Resistance to Severe Ethanol Stress in Saccharomyces cerevisiae Protein Quality Control: Yoshida, M., Kato, S., et al.; Appl. Environ. Microbiol. 87, (2021), Abstract
- Cytosolic aggregation of mitochondrial proteins disrupts cellular homeostasis by stimulating the aggregation of other proteins: Vendruscolo, M., Chacinska, A., et al.; bioRxiv , (2021), Reactant(s): Saccharomyces cerevisiae
- Cytosolic aggregation of mitochondrial proteins disrupts cellular homeostasis by stimulating the aggregation of other proteins.: Dobson, C. M., Vendruscolo, M., et al.; Elife 10, (2021), Application(s): WB / Reactant(s): Saccharomyces cerevisiae, Abstract
- Functional analysis of proposed substrate-binding residues of Hsp104: M.K. Howard, et al.; PLoS One 15, e0230198 (2020), Abstract — Full Text
- Chaperone-Mediated Protein Disaggregation Triggers Proteolytic Clearance of Intra-nuclear Protein Inclusions.: den Brave, F., Cairo, L. V., et al.; Cell Rep. 31, 107680 (2020), Reactant(s): Saccharomyces cerevisiae, Abstract
- Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness.: Tye, B. W., Commins, N., et al.; Elife 8, (2019), Application(s): WB / Reactant(s): Saccharomyces cerevisiae, Abstract
- Mining Disaggregase Sequence Space to Safely Counter TDP-43, FUS, and α-Synuclein Proteotoxicity: Tariq, A., Lin, J., et al.; Cell Rep. 28, 2080 (2019), Abstract
- Engineered protein disaggregases mitigate toxicity of aberrant prion-like fusion proteins underlying sarcoma: J.J. Ryan, et al.; J. Biol. Chem. 294, 11296 (2019), Abstract
- A risk-reward tradeoff of high ribosome production in proliferating cells: Springer, M., Pincus, D., et al.; bioRxiv , (2018), Application(s): WB / Reactant(s): Saccharomyces cerevisiae
- FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination: K. Yasuda, et al.; J. Cell Biol. 216, 1015 (2017), Application(s): WB / Reactant(s) Mouse, Abstract — Full Text
- Mechanistic Insights into Hsp104 Potentiation: M.P. Torrente, et al.; J. Biol. Chem. 291, 5101 (2016), Application(s): WB / Reactant(s) Saccharomyces cerevisiae, Abstract — Full Text
- Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol.: Bragoszewski, P., Oeljeklaus, S., et al.; Nature 524, 485 (2015), Reactant(s): Saccharomyces cerevisiae, Abstract
- Cytosolic chaperones mediate quality control of higher-order septin assembly in budding yeast.: Thorner, J., Weems, A. D., et al.; Mol. Biol. Cell 26, 1323 (2015), Reactant(s): Saccharomyces cerevisiae, Abstract
- Hsp104 suppresses polyglutamine-induced degeneration post onset in a drosophila MJD/SCA3 model.: Bonini, N. M., Shorter, J., et al.; PLoS Genet. 9, e1003781 (2013), Application(s): IHC-Frozen, WB / Reactant(s): Drosophila melanogaster, Abstract
- In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other: E. Cabib, et al.; J. Cell. Sci. 115, 293 (2002), Application(s): WB using yeast samples, Abstract
- A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress: D.C. Masison, et al.; Genetics 156, 559 (2000), Application(s): WB using yeast samples, Abstract
- Prion-dependent switching between respiratory competence and deficiency in the yeast nam9-1 mutant: S. Rospert, et al.; Mol. Cell. Biol. 20, 7220 (2000), Application(s): WB using yeast samples, Abstract
- Hsp104 is a highly conserved protein with two essential nucleotide-binding sites: S. Lindquist, et al.; Nature 353, 270 (1991), Application(s): WB using yeast samples, Abstract
Related Products
| Application | ELISA, IHC, WB |
|---|---|
| Host | Goat |
| Species Reactivity | Rabbit |
Last modified: May 29, 2024
Datasheet, Manuals, SDS & CofA
Manuals And Inserts
Certificate of Analysis
Please enter the lot number as featured on the product label
SDS
Enzo Life Science provides GHS Compliant SDS
If your language is not available please fill out the SDS request form
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?
