Contamination Control of Genomic DNA

gDNA Removal Kit
Remove contaminating gDNA for more reliable data
Contaminating genomic DNA (gDNA) in RNA preparations can lead to incorrect quantification of RNA in RT-PCR or high DNA background in RNA sequencing. This is particularly an issue during the quantification of low-copy transcripts or the sequencing of small RNA samples. Removal of gDNA is often a necessity to prevent high DNA background in RNA sequencing. This protocol can be applied to RNA preparations before next-generation sequencing (NGS).
The gDNA removal kit is based on a recombinant heat-labile dsDNase (HL-dsDNase) to remove gDNA from RNA preparations. Through HL-dsDNase technology, contaminating genomic DNA is cleaved at ambient temperature and the enzyme is irreversibly inactivated upon increasing the temperature over 50° C.
Complete removal of gDNA from RNA extractions
dsDNase inactivated without reducing RNA quality or quantity
Simple
Minimizes pipetting steps and reduces hands-on time
Scalable
Suitable for high-throughput experiments
Fast
Eliminate gDNA contamination in RNA samples within 20 minutes
It’s as easy as Mix, Heat, and Run
Complete Removal of gDNA from RNA Isolation
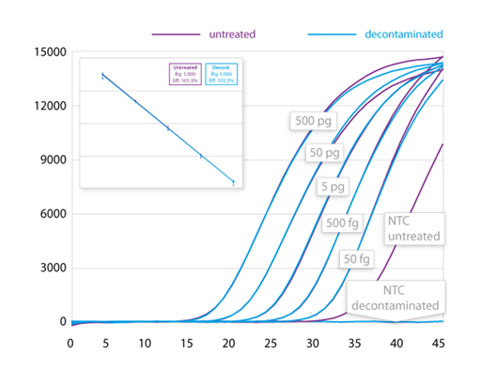
The gDNA is removed from RNA preparations to levels below the detection limit of RT-qPCR and the inactivation is gentle enough to preserve both quality and quantity of all present RNA (Fig. 1A and B).
References
N. Vollebergh, M.F. Alexeyev. Limited predictive value of TFAM in mitochondrial biogenesis. Mitochondrian (2019), doi: 10.1016/j.mito.2019.08.001
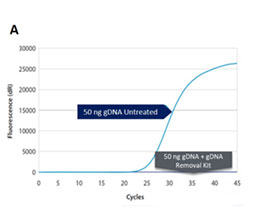
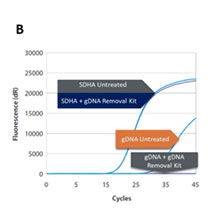
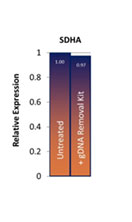
Removal of gDNA in RNA preparations. (A) The gDNA removal kit removes at least 50 ng of gDNA in a 10 µL reaction volume. (B) Compared to the untreated control, no reduction in quantified cDNA (SDHA) was detected
PCR Decontamination Kit
Get control of your contamination
Rapid removal of contaminating DNA in PCR mastermixes, without reduction in PCR sensitivity
PCR is a sensitive technique used in research and diagnostics. Taq polymerases, dNTP’s, buffer components, and primers/probes are frequently contaminated with E. coli DNA, which may lead to false positives if bacterial DNA is the target.
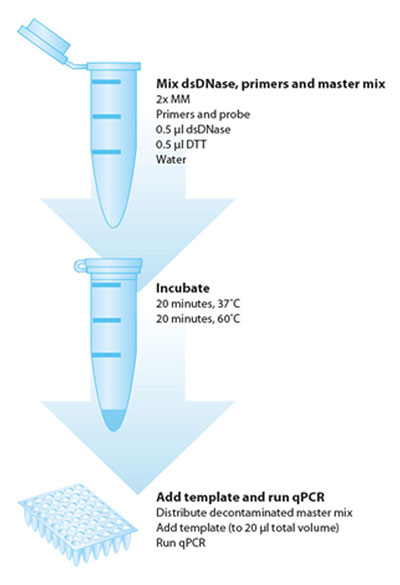
The PCR decontamination kit uses a dsDNase to remove contaminating DNA from mastermixes. The double-strand specific property allows decontamination in the presence of primers and probe. The dsDNase is irreversibly inactivated by heating to 60°C in the presence of DTT, ensuring that DNA template added to the mastermix after inactivation will not be digested. The mastermix can be used immediately after decontamination to run PCR reactions.
AMPIGENE® Taq polymerases have been treated to remove host DNA and additional clean-up is not necessary. The PCR decontamination kit is recommended for decontamination of primers or competitor’s PCR mixes.
Sample decontamination workflow
PCR & Gel Clean-up Columns
Quick and simple kit for purifying DNA from PCR, gels, and labeling reactions for a variety of downstream applications. Silica membrane technology in a mini spin column format to deliver reliable DNA purification from PCR products, agarose gels, and labeling reactions.
High performance
Recover up to 95% of DNA for fragments down to 50 bp
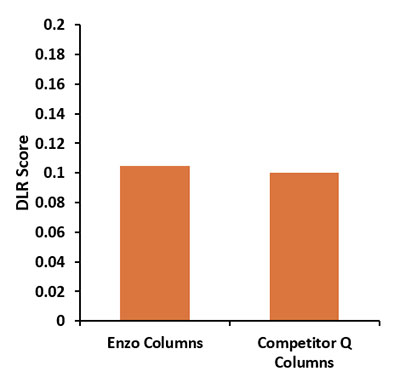
Enzo’s Columns Perform Equivalently to Leading Competitor’s Columns Post-CGH Labeling
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?