BIOARRAY™ Amplification & Labeling Systems
Optimized for routine GeneChip® analysis
Enzo’s BIOARRAY™ RNA Amplification & Labeling kits offer labeling solutions for biotin-based gene expression microarrays. Due to greater biotin incorporation and brighter signal, BIOARRAY™ RNA labeling is the best choice for routine GeneChip® analysis.
BIOARRAY™ Single-round RNA amplification and biotin labeling system
Improved data quality and array analysis through greater biotin incorporation:
Biotin-labeled antisense RNA (aRNA) is generated from total cellular RNA samples in less than 24 hours. Incorporation of two biotin nucleotides yields brighter signal, improving data from microarray experiments.
- Convenient workflow with a flexible 4-16 hour transcription time and reagents supplied in a ready-to-use format.
- Decrease experimental variation, and standardize data derived from microarrays.
- Maintain the value of legacy data by the continued use of the gold standard for GeneChip® visualization.
- Enables correlation of results from experiment-to-experiment, project-to-project and lab-to-lab.
- Production of large amounts of biotin-labeled RNA targets by in vitro transcription from bacteriophage T7 RNA polymerase promoters is available separately with our BioArray HighYield® RNA Transcript Labeling Kits.
…our results indicate that
the Enzo kit is the best choice for routine Affymetrix GeneChip experiments.
Read the complete Application Note
References
- Rosen, MB et.al. Gene Expression Profiling in Wild-Type and PPARα-Null Mice Exposed to Perfluorooctane Sulfonate Reveals PPARα-Independent Effects. PPAR Research Volume 2010, Article ID 794739, doi:10.1155/2010/794739.
- Gabelt, BT et.al. Caldesmon effects on the actin cytoskeleton and cell adhesion in cultures HTM cells. Experimental Eye Research. Volume 82, issue 6, June 2006, 945-958.
Analyze Limiting Quantities of Total RNA
Analyze limiting quantities of total RNA with the BIOARRAY™ Low Input RNA Amplification and Biotin Labeling System for transcript analysis
- Generates sufficient aRNA for standard microarray analysis from as little as 20 ng of total input RNA.
- Entire amplification reaction can be performed in a single tube.
- Superior 3’/5’ transcript ratios demonstrating efficient in vitro transcription.
- Biotin-labeled aRNA can be purified using either magnetic beads or purification columns and reagents
BIOARRAY™ 3´-OH Terminal Labeling is the recognized benchmark standard for biotin-labeling of DNA
The kit is considered the gold standard end-labeling system for use with Affymetrix® DNA SNP (single nucleotide polymorphism), resequencing and prokaryotic microarrays. This method uses Bio-ddUTP and terminal deoxynucleotide transferase to catalyze the addition of a single biotin-ddUMP (2’,3’-dideoxyuridine 5’-monophosphate) to the 3’-OH terminus of an amplified and fragmented target DNA molecule.
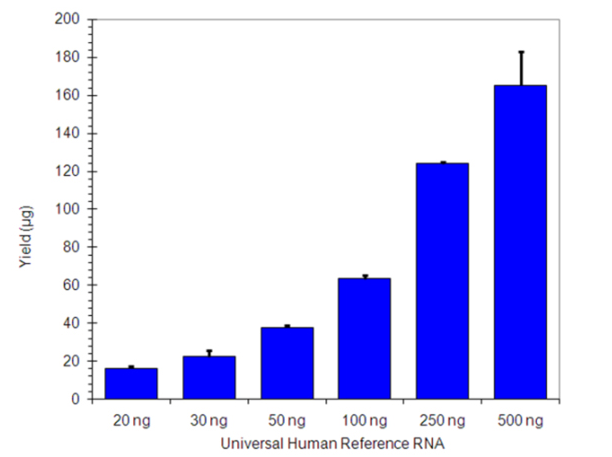
As little as 20 ng total RNA input produces sufficient amounts of aRNA for microarrays
Figure 1: Universal Human Reference RNA ranging from 20ng to 500ng was amplified in triplicate using BIOARRAY™ Low Input RNA Amplification and Biotin Labeling System. The lowest amount of input (20ng) generated enough labeled aRNA for microarray analysis.
References
Akiyama, M et.al. Analysis of telomerase activity and RNA expression in a patient with acute promyelocytic leukemia treated with all-trans retinoic acid. Pediatric Blood & Cancer. Volume 46, Issue 4, 2006, 506-511.
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?
