Jack Coleman1, Wini Luty1, Marie-Laure Schneider2
1Enzo Life Sciences, Farmingdale, NY USA
2Innopsys, Toulouse, France
INTRODUCTION
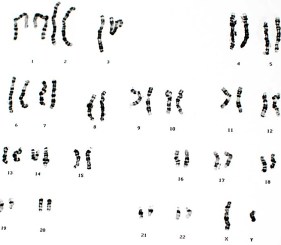
Array CGH is a well-established method for investigating copy number variations. It is used to detect a variety of genomic gains and losses ranging from copy number variants, duplications and deletions, unbalanced translocations, and aneuploidies.
In an array CGH experiment, test DNA and reference DNA are labeled with two different fluorophores. Equal amounts are combined and hybridized to a microarray containing DNA or oligonucleotide probes representing small sections of chromosomes. This holistic approach allows the rapid screening of the entire genome while also enabling the investigation of changes in sequence of a particular section of a chromosome.
In addition, single nucleotide polymorphism (SNP), variation at a single nucleotide in a genome, can also be examined on a CGH+SNP array. A CGH+SNP array enables detection of genomic aberrations associated with copy number variations, but in addition, it can also detect copy neutral aberrations, such as loss of heterozygosity (LOH) and uniparental disomy (UPD), on the same array.
The main objective of this study was to demonstrate the superior labeling capability of the CYTAG™ TotalCGH Labeling Kit in preparing and labeling DNA for a CGH+SNP microarray, and further, to analyze the chip on the Innopsys InnoScan® 910.
Download this Application Note
Latest Articles



 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?