Idrissia Hanriot1 | Cécile Odin1 | Jessica Demange1 | Rosaria Esposito2 | Morgan Mathieu3 | Jean-Louis Merlin1
- Tumor Molecular Biology Service / Department of Biopathology, CHRU-ICL Nancy
- Enzo Life Sciences, Villeurbanne, France
- Enzo Life Sciences, Lausen, Switzerland
INTRODUCTION
Microarray-based comparative genomic hybridization (array CGH) is a fast genome-wide screening tool for the high-resolution detection of a large number of chromosomal anomalies, both constitutional and acquired, in the form of copy number variations (CNVs), such as duplications and deletions, unbalanced translocations, and aneuploidies. This relatively simple technology has been used extensively in clinical settings, including prenatal and postnatal diagnostics.
Single nucleotide polymorphism (SNP) (i.e., variation at a single nucleotide in a genome) can be examined on a CGH+SNP array. A CGH+SNP array enables the detection of genomic aberrations associated with copy number variations.
It can also detect copy neutral aberrations on the same array, such as loss of heterozygosity (LOH) and uniparental disomy (UPD).
In the oncology field, array CGH represents a valuable prognostic tool. It allows the identification of the genetic alterations characterizing a specific patient’s tumor, giving precious information on its evolution, and
facilitating the choice of the best treatment. However, biopsies are usually preserved in a formalin-fixed paraffin-embedded (FFPE) form,
or by freezing the specimen. Both the quantity and the quality of the DNA extracted from these tissues are variable, making the analysis by CGH of these specimens challenging.
Download this Application Note
Benefits
- CYTAG® SuperCGH+SNP labeling kit yields lower DLR scores, making the plots easier to interpret
- Provides dynamic analytical range for
challenging FFPE and frozen oncological tissues - Easy and fast protocol
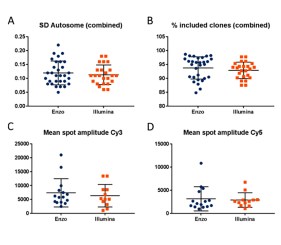
This is reflected in the high derivative log ratio (DLR) scores often obtained with oncological samples. The DLR score is a measurement of point-to-point consistency, and is one of the parameters commonly used to evaluate the quality of a CGH profile. Low DLR scores reduce the need for experimental repeats and are key to interpreting CGH data. In general, DLR scores should be lower than 0.20 to be considered optimal, but when dealing with oncological samples, even DLR scores lower than 0.30 are often difficult to achieve. Interpreting SNP profiles with such complex input material is also often extremely challenging, and clean results are pivotal for correctly interpreting the data.
The Tumor Molecular Biology Service at the CHRU-ICL Nancy carries out hundreds of oncological CGH tests per year on FFPE and frozen tissues. The objective of this study was to test the performance of the new Enzo Life Sciences’ CYTAG® SuperCGH+SNP Labeling Kit with DNA extracted from oncological tissues (FFPE or frozen), and to compare the results with previous data obtained by labeling the same samples with the kit from a competitor routinely used in the laboratory.
Latest Articles




 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?