Jack Coleman1, Wini Luty1, Marie-Laure Schneider2
1Enzo Life Sciences, Farmingdale, NY USA
2Innopsys, Toulouse, France
INTRODUCTION
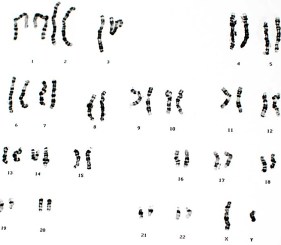
Array-based comparative genomic hybridization (aCGH) is a powerful tool for detecting copy number variations. It is a well-established method used to analyze genomic gains and losses ranging from duplications, deletions, unbalanced translocations, and aneuploidies.
Array CGH is a universal approach that enables the rapid screening of the entire genome. Test DNA and reference DNA are labeled with two different fluorophores and equal amounts are hybridized to a microarray. The microarrays contain either DNA or oligonucleotide probes representing small sections of chromosomes. Array CGH looks at chromosomal imbalances between the test DNA and the reference DNA and is capable of giving a holistic comparison of the genome.
The main objective of this study was to utilize the high resolution Innopsys InnoScan 910 to demonstrate the superior performance of the CYTAG™ CGH Labeling Kit in labeling DNA for CGH analysis on a 4×180 whole genome microarray.
Download this Application Note
Latest Articles



 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?