Brad Larson1, Leonie Rieger1, Glauco Souza2, Hubert Tseng2, Arun Kumar3
1 BioTek Instruments, Inc., Winooski, Vermont, USA
2 Nano3D Biosciences, Inc., Houston, Texas, USA
3 Enzo Life Sciences, Farmingdale, New York, USA
INTRODUCTION
Colorectal cancer (CRC) is one of the most common cancer types in the U.S., with the National Cancer Institute estimating almost 1.2 million people nationwide living with the disease as of 2013, and over 135,000 new cases expected in 20171. For patients with advanced stages of CRC, there is a meager survival rate due to the cancer’s predisposition to metastasize and resist treatment therapies. Previous studies have shown that the two complexes of the serine/threonine kinase mammalian target of rapamycin (mTOR), mTORC1 (containing Raptor) and mTORC2 (containing Rictor) are overexpressed in CRC and play a role in tumorigenesis and metastasis2, 3. Furthermore, upregulation of the mTOR signaling pathway has also been shown to induce expression of matrix metalloproteinases (MMPs), which also play a role in CRC metastagenicity4. Therefore, development of therapies that incorporate mTOR inhibitors may reduce the metastatic potential of this cancer and improve survival rates.
Download this Application Note
As colorectal cancers are solid tumors that grow in vivo in a three dimensional (3D) configuration, a major limitation in the vast number of studies involving CRC research is the use of cells cultured onto a two-dimensional (2D) surface. Furthermore, cell metastatic behavior is often studied in vitro using phenotypic cell migration and target-based mechanism of action models. In regards to the presented study, mTOR signaling is known to play a role in cell migration and cell-extracellular matrix (ECM) formation, which also leads to downstream expression of MMPs. The use of a 2D solid surface environment common to traditional labware results in altered cellular morphologies and behavior, as well as a lack of complex cell-cell or ECM interaction networks which often influences cell function, ECM production, and cell migration. 3D cell cultures remedy these shortcomings by providing a method that enables cells to reorganize and migrate into a structure that better replicates an in vivo microenvironment by capturing cell-cell and cell ECM interactions. This development provides an improved predictive in vitro model for cancer metastasis.
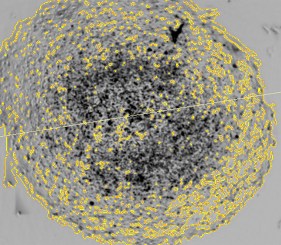
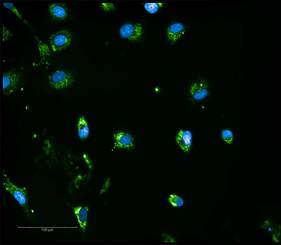
Here, we demonstrate a magnetic 3D bioprinting cell culture platform (Figure 1), where cells are first incubated with a biocompatible magnetic nanoparticle assembly consisting of gold, iron oxide and poly-L-lysine, which magnetizes the cells without eliciting deleterious biological effect. The cells are then placed into a microplate well and levitated by placing a magnet above the well, where they aggregate and form ECM within a few hours. After this initial levitation step, the magnet is removed, and the 3D aggregates are dissociated into a disperse cell suspension of single cells and small cell aggregates by gentle pipetting action. Cells are then transferred to a 384-well assay plate and a spheroid magnet is positioned below the plate for an appropriate incubation period, allowing the cells within each well to be patterned into a dot or spheroid configuration depending upon the experiment being performed. Moreover, the magnetized spheroid can be held intact while exchanging liquids to improve sample retention, such as during regular media exchanges. Following removal of the magnet, protein expression within spheroid models was quantified via immunofluorescence, while rates of phenotypic cell migration were assessed using kinetic live cell brightfield imaging to track cell and ECM movement away from the original pattern. Mechanism of action and tumorigenesis were also determined by microplate reading and fluorescent imaging, respectively. Normal and cancerous co-cultured colon cell models were incorporated to test protein expression, whereas cell migration was analyzed using HCT116 colorectal cancer cells only. Automated imaging and microplate reading were performed using a novel cell imaging multi-mode reader. The combination of 3D cell model and image-based analysis provides easy-to-use, robust methods to quantify protein expression and 3D cell migration allowing confirmation of the role that mTOR signaling plays in the development and treatment of colorectal cancer.
Latest Articles
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?