Andrew Lithen, Shay Karkashon
Enzo Life Sciences, Farmingdale, NY USA
INTRODUCTION
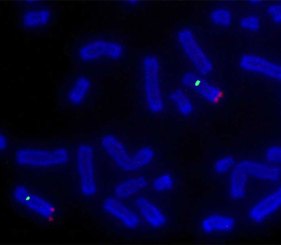
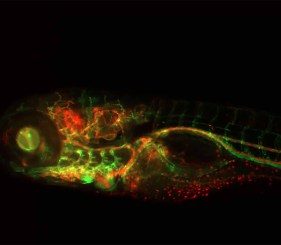
Labeling DNA with fluorescent dye dUTPs is well recognized as superior to labeling DNA with indirect two-step labeling methods. Coupled with the Nick Translation DNA Labeling System 2.0, fluorescent dUTPs provide a simple and efficient approach to fluorescent labeling for FISH applications.
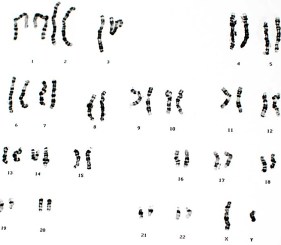
Accommodating a wide range of fluorophore-labeled, biotin-labeled, and digoxigenin-labeled nucleotides, the Nick Translation DNA Labeling System 2.0 is the recommended method for labeling double-stranded DNA larger than 1 kb for applications including fluorescent in situ hybridization (FISH). FISH is a well-documented technique used to label DNA sequences in chromosomes for research and clinical applications.
Relatively humid environmental conditions are necessary for proper fluorescent in situ hybridization; however, the optimal humidity for FISH is not yet known. The main objective of this study was to demonstrate the capabilities of the Nick Translation System 2.0 for FISH under varying levels of humidity with the Boekel Scientific RapidFISH Slide Hybridization Oven.
 Lab Essentials
Lab Essentials AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?