Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
This antibody is covered by our Worry-Free Guarantee.
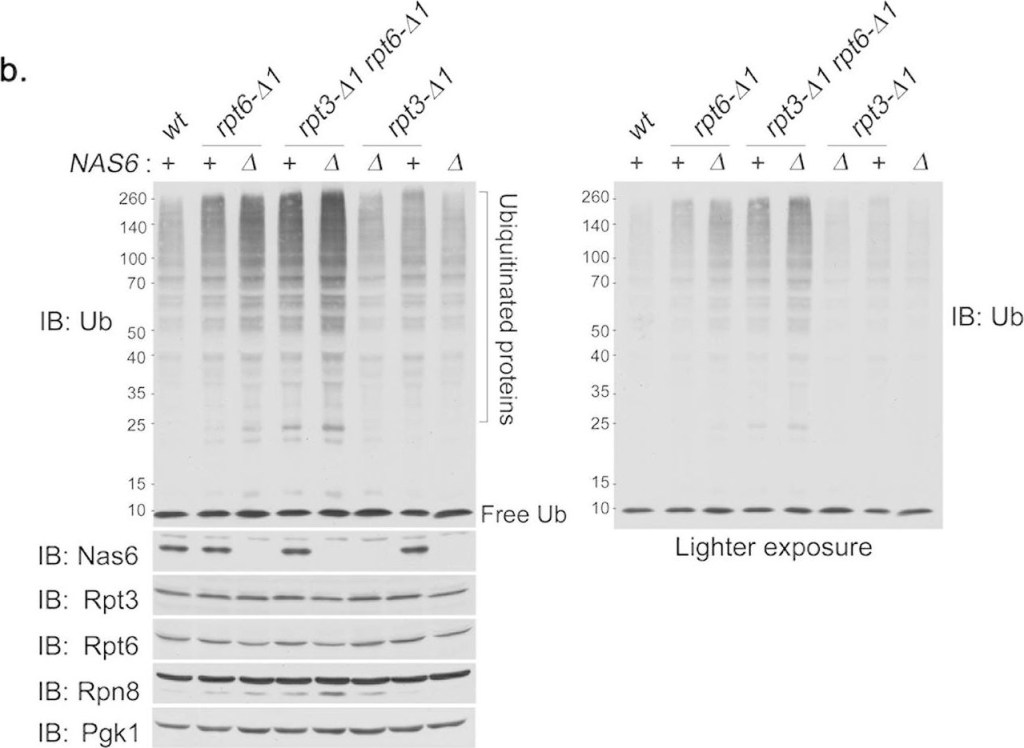
The Rpt6 tail exhibits a distinct functional relationship with Nas6 in vivo.(a) Phenotypic analysis showing effect of nas6Δ on the growth of rpt6-Δ1 or rpt3-Δ1 single, or double mutants. Four-fold serial dilutions of indicated cells were spotted onto YPD plates, synthetic complete medium (SC), or SC medium containing canavanine (1 μg/ml), and incubated for 2–3 days at the indicated temperature. For testing sensitivity to canavanine (an arginine analog), arginine was omitted from the SC medium. (b) Effect of nas6Δ on the degradation of polyubiquitinated proteins in rpt6-Δ1 or rpt3-Δ1 single, or double mutant cells. The cells were cultured for 6 hours at 37 °C. Whole cell lysates (20 μg) were subjected to 10% Bis-Tris SDS-PAGE for immunoblotting (IB) of polyubiquitinated proteins, and 12% SDS-PAGE for immunoblotting of Nas6 and proteasome subunits. Rpt3 and Rpt6 are base subunits. Rpn8 is a lid subunit. Pgk1 serves as a loading control. Lighter exposure of anti-ubiquitin (Ub) immunoblot is shown at right to further illustrate the difference in polyubiquitinated protein levels. Molecular weight markers are at left in kDa.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep (2015)
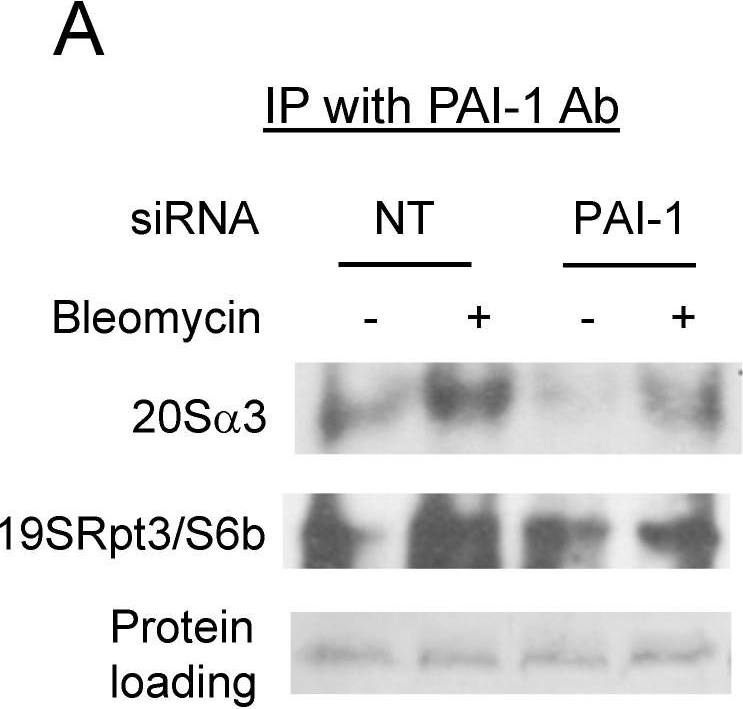
PAI-1 binds to proteasome components in A549 cells. (A,D) Immunoprecipitation-immunoblotting analysis of PAI-1 interaction with proteasome 20S α3 and 19S Rpt3/S6b subunits in A549 cells. A549 cells were transfected with PAI-1 siRNA/non-target (NT) siRNA or transduced with control/PAI-1 expressing viruses and then treated with bleomycin. PAI-1 protein was immunoprecipitated with anti-mouse PAI-1 monoclonal antibody, and Westerns were conducted with specific antibody to 20Sα3 or 19SRpt3/S6b. Proteins on the membrane were stained with Ponceau S to show equal sample loading. (B,C,E,F) Semi-quantification of the band intensities, normalized by the corresponding protein staining band (n = 3).
Image collected and cropped by CiteAb under a CC-BY license from the following publication: PAI-1 Regulation of p53 Expression and Senescence in Type II Alveolar Epithelial Cells. Cells (2023)
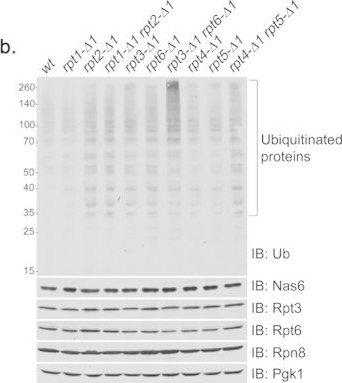
Together, the Rpt3 and Rpt6 tails play a central role in proteasome function.(a) Yeast growth assay showing severe heat sensitivity of rpt3-Δ1rpt6-Δ1 double mutant cells. Four-fold serial dilutions of indicated yeast strains were spotted onto YPD plates and grown for 2–3 days at 30 °C and 37 °C. Table 1 lists the yeast strains used in each figure henceforth. (b) Anti-ubiquitin immunoblots showing an accumulation of polyubiquitinated proteins in rpt3-Δ1rpt6-Δ1 cells. Levels of the Nas6 chaperone and proteasome subunits remained largely unchanged in all indicated strains. Whole cell lysates (20 μg) were subjected to 10% Bis-Tris SDS-PAGE for immunoblotting (IB) of polyubiquitinated proteins, and 12% Tris-Glycine SDS-PAGE (SDS-PAGE henceforth) for immunoblotting of Nas6 and proteasome subunits. Nas6 is a cognate chaperone of Rpt3. Rpt3 and Rpt6 are base subunits. Rpn8 is a lid subunit. Pgk1 serves as a loading control. Ub is ubiquitin. Molecular weight markers are at left in kDa.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep (2015)
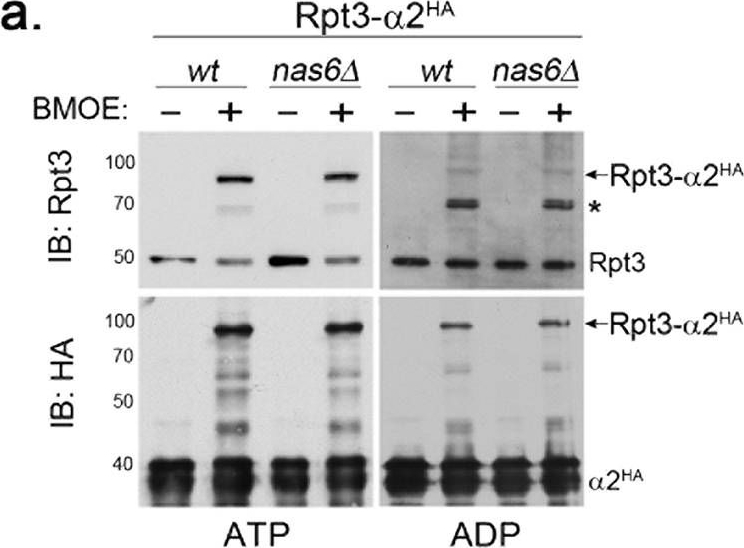
The Rpt6 tail-α3 interaction decreases in the proteasome holoenzyme and is less nucleotide-dependent than the Rpt3 tail-α2 interaction.(a) Rpt3 tail-α2 binding is strongly ATP-dependent in the proteasome holoenzyme, but is Nas6-independent. Wild-type and nas6Δ cells each harbor rpt3-K428C and α2-A79C-HA6 alleles; K428 is the last residue of Rpt3. Proteasomes were immunoprecipitated via α2-HA6 in the presence of ATP (1 mM) or ADP (2 mM), and then incubated with a chemical crosslinker BMOE (0.1 mM), or its solvent, DMF for 1 hour at 4 °C and subjected to SDS-PAGE. Rpt3-α2HA crosslinks were detected by immunoblotting (IB) for Rpt3 and α2HA. Molecular weight markers are at left in kDa. Asterisk (*) indicates a non-specific signal. Crosslinked products remain stable during our analysis since BMOE is an irreversible crosslinker. (b) Rpt6 tail-α3 binding occurs in both ATP and ADP in the proteasome holoenzyme. Experiments were conducted as in (a) in wild-type and nas6Δ cells, each harboring rpt6-K405C and α3-T81C-HA6 alleles; K405 is the last residue of Rpt6. Rpt6-α3HA crosslinks were detected by immunoblotting for Rpt6 and α3HA. Asterisk (*) indicates a non-specific band. (c) Rpt6-α3 interaction decreases in the proteasome holoenzyme whereas Rpt3-α2 interaction increases. The Rpt3-α2 crosslinks and Rpt6-α3 crosslinks as in (a,b) were quantified using ImageJ software and shown as mean + SEM (n = 6, Rpt3-α2, ATP; n = 3, Rpt3-α2, ADP and Rpt6-α3, ATP; n = 4, Rpt6-α3, ADP). The ratio on the Y axis was obtained by normalizing the intensities of Rpt-αHA crosslinked bands to corresponding uncrosslinked Rpt bands on the same immunoblot, for example, [Rpt6-α3HA band (80 kDa)]/[Rpt6 band (45 kDa)] (Fig. 6b, lane 2, top). Note that 43.6 + 0.6% decrease in Rpt6-α3HA ratio (ATP) is relative to Rpt3-α2HA ratio (ATP). (d) Rpt6-α3 interaction occurs in ADP whereas Rpt3-α2 interaction severely decreases. The Y axis indicates percent decrease of Rpt-αHA ADP samples relative to their corresponding ATP samples from (c). (e) The Rpt6 tail is crucial for Rpt3-α2 binding (arrow heads) in the proteasome holoenzyme. Experiments were conducted as in (a) in rpt6-Δ1 and wild-type cells, each carrying rpt3-K428C and α2-A79C-HA6 alleles. Rpt6 levels remain unchanged.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep (2015)
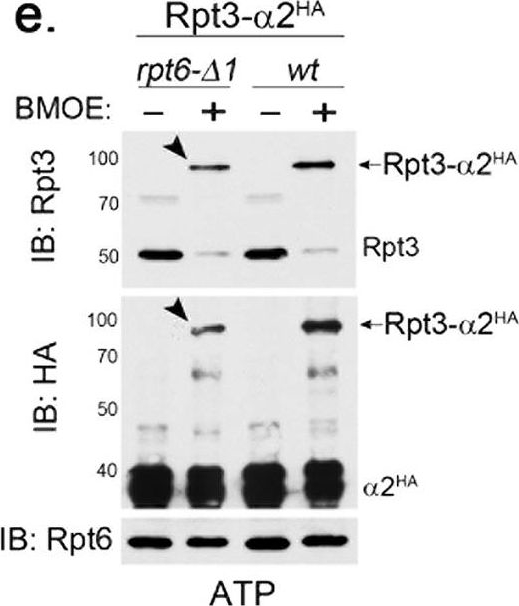
The Rpt6 tail-α3 interaction decreases in the proteasome holoenzyme and is less nucleotide-dependent than the Rpt3 tail-α2 interaction.(a) Rpt3 tail-α2 binding is strongly ATP-dependent in the proteasome holoenzyme, but is Nas6-independent. Wild-type and nas6Δ cells each harbor rpt3-K428C and α2-A79C-HA6 alleles; K428 is the last residue of Rpt3. Proteasomes were immunoprecipitated via α2-HA6 in the presence of ATP (1 mM) or ADP (2 mM), and then incubated with a chemical crosslinker BMOE (0.1 mM), or its solvent, DMF for 1 hour at 4 °C and subjected to SDS-PAGE. Rpt3-α2HA crosslinks were detected by immunoblotting (IB) for Rpt3 and α2HA. Molecular weight markers are at left in kDa. Asterisk (*) indicates a non-specific signal. Crosslinked products remain stable during our analysis since BMOE is an irreversible crosslinker. (b) Rpt6 tail-α3 binding occurs in both ATP and ADP in the proteasome holoenzyme. Experiments were conducted as in (a) in wild-type and nas6Δ cells, each harboring rpt6-K405C and α3-T81C-HA6 alleles; K405 is the last residue of Rpt6. Rpt6-α3HA crosslinks were detected by immunoblotting for Rpt6 and α3HA. Asterisk (*) indicates a non-specific band. (c) Rpt6-α3 interaction decreases in the proteasome holoenzyme whereas Rpt3-α2 interaction increases. The Rpt3-α2 crosslinks and Rpt6-α3 crosslinks as in (a,b) were quantified using ImageJ software and shown as mean + SEM (n = 6, Rpt3-α2, ATP; n = 3, Rpt3-α2, ADP and Rpt6-α3, ATP; n = 4, Rpt6-α3, ADP). The ratio on the Y axis was obtained by normalizing the intensities of Rpt-αHA crosslinked bands to corresponding uncrosslinked Rpt bands on the same immunoblot, for example, [Rpt6-α3HA band (80 kDa)]/[Rpt6 band (45 kDa)] (Fig. 6b, lane 2, top). Note that 43.6 + 0.6% decrease in Rpt6-α3HA ratio (ATP) is relative to Rpt3-α2HA ratio (ATP). (d) Rpt6-α3 interaction occurs in ADP whereas Rpt3-α2 interaction severely decreases. The Y axis indicates percent decrease of Rpt-αHA ADP samples relative to their corresponding ATP samples from (c). (e) The Rpt6 tail is crucial for Rpt3-α2 binding (arrow heads) in the proteasome holoenzyme. Experiments were conducted as in (a) in rpt6-Δ1 and wild-type cells, each carrying rpt3-K428C and α2-A79C-HA6 alleles. Rpt6 levels remain unchanged.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep (2015)
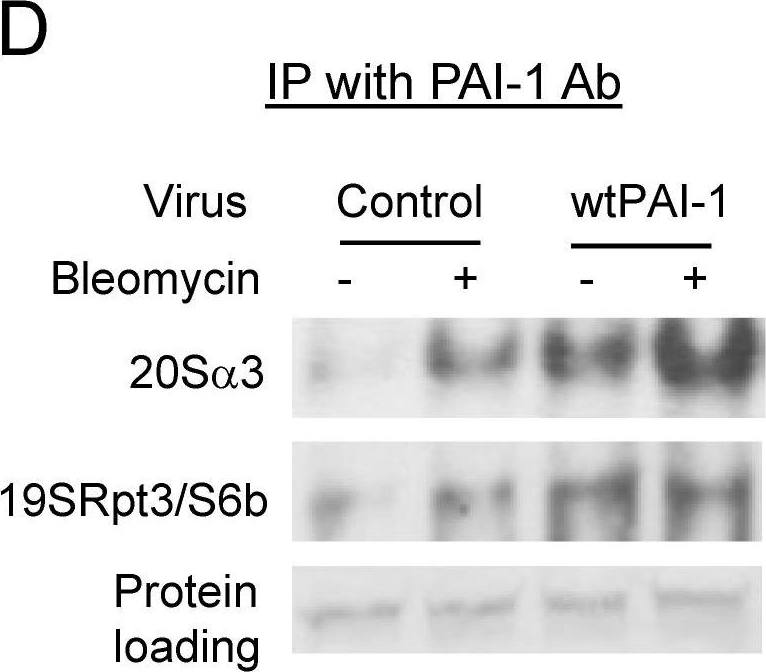
PAI-1 binds to proteasome components in A549 cells. (A,D) Immunoprecipitation-immunoblotting analysis of PAI-1 interaction with proteasome 20S α3 and 19S Rpt3/S6b subunits in A549 cells. A549 cells were transfected with PAI-1 siRNA/non-target (NT) siRNA or transduced with control/PAI-1 expressing viruses and then treated with bleomycin. PAI-1 protein was immunoprecipitated with anti-mouse PAI-1 monoclonal antibody, and Westerns were conducted with specific antibody to 20Sα3 or 19SRpt3/S6b. Proteins on the membrane were stained with Ponceau S to show equal sample loading. (B,C,E,F) Semi-quantification of the band intensities, normalized by the corresponding protein staining band (n = 3).
Image collected and cropped by CiteAb under a CC-BY license from the following publication: PAI-1 Regulation of p53 Expression and Senescence in Type II Alveolar Epithelial Cells. Cells (2023)






Product Details
| Alternative Name |
26S protease regulatory subunit 6B, Tat-binding protein 7, TBP7, Proteasome 26S subunit ATPase 4 |
|---|---|
| Application |
IHC, WB |
| Formulation |
Liquid. In PBS containing 10mM sodium azide. |
| GenBank ID |
464861 (S. cerevisiae) |
| Host |
Rabbit |
| Immunogen |
Recombinant protein corresponding to the N-terminal 100 amino acids of the YTA2 protein. |
| Species Reactivity |
Human, Yeast |
| Specificity |
Recognizes the Rpt3/S6b subunit of the 19S regulator complex. |
| UniProt ID |
P33298 (S. cerevisiae), P43686 (human) |
| Worry-free Guarantee |
This antibody is covered by our Worry-Free Guarantee. |
Handling & Storage
| Long Term Storage |
-20°C |
|---|---|
| Shipping |
Blue Ice |
| Regulatory Status |
RUO – Research Use Only |
|---|
- PAI-1 Regulation of p53 Expression and Senescence in Type II Alveolar Epithelial Cells.: Rana, T., Jiang, C., et al.; Cells 12, (2023), Application(s): WB / Reactant(s): Human, Abstract
- Assembly checkpoint of the proteasome regulatory particle is activated by coordinated actions of proteasomal ATPase chaperones: A. Nahar, et al.; Cell Rep. 39, 110918 (2022), Abstract
- Homeostatic scaling is driven by a translation-dependent degradation axis that recruits miRISC remodeling: Srinivasan, B., Samaddar, S., et al.; PLoS Biol. 19, e3001432 (2021), Abstract
- Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly: V. Sokolova, et al.; Sci. Rep. 5, 14909 (2015), Application(s): WB / Reactant(s) Saccharomyces cerevisiae, Abstract — Full Text
Datasheet, Manuals, SDS & CofA
Manuals And Inserts
Certificate of Analysis
Please enter the lot number as featured on the product label
SDS
Enzo Life Science provides GHS Compliant SDS
If your language is not available please fill out the SDS request form
 AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?
