Shipping: Available products typically ship within 24/48h, via priority shipping.
Do you need support? Contact Customer Service or Technical Support.
Online Account
Access or Create Your Account
This antibody is covered by our Worry-Free Guarantee.
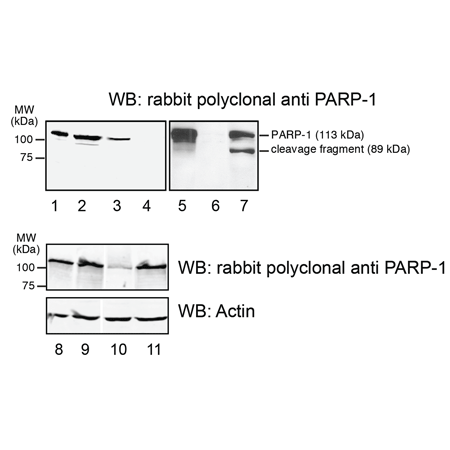
Figure: Western blot using rabbit polyclonal anti PARP-1.
Lane 1: recombinant human PARP-1 (ALX-201-053, 5 ng).
Lane 2: Total HeLa cell extract.
Lane 3: Total MEF PARP1+/+ cell extract.
Lane 4: Total MEF PARP1-/- cell extract.
Lane 5: Lysate (50 μg) from HeLa cells.
Lane 6: Lysate (50 µg) from HeLa PARP-1sh cells.
Lane 7: Lysate (50 µg) from HeLa cells treated for 8 hours with Doxorubicin 5 µg/ml.
Lane 8: Lysate (50 µg) from HEK293 cells.
Lane 9: Lysate (50 µg) from HEK293 cells.
Lane 10: Lysate (50 µg) from HEK293 cells transfected with PARP-1 siRNA.
Lane 11: Lysate (50 μg) from HEK293 cells transfected with control siRNA.
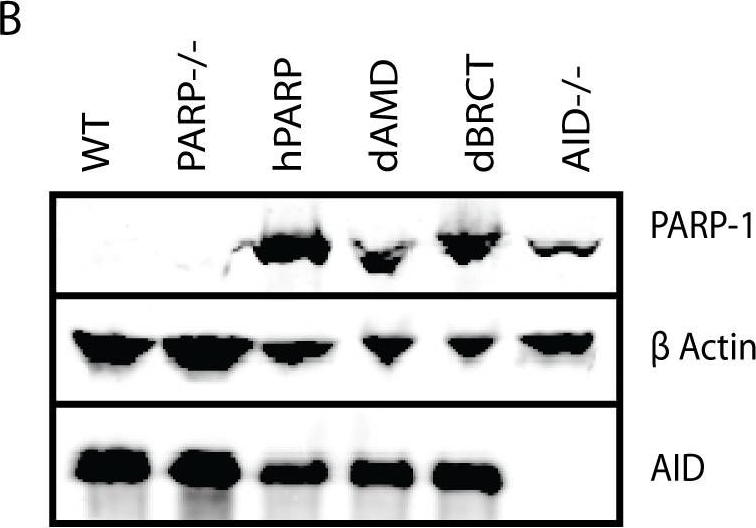
The PARP-1 BRCT domain is required for immunoglobulin gene conversion.(A) Schematic of the domains of PARP-1 and variants dAMD and dBRCT. (B) Western blot showing levels of PARP-1 and AID expression with β actin as a loading control. (C) Survival of PARP-1 variants to MMS-induced DNA damage. Experiment was performed in triplicate and error bars represent SEM. *** p<.0001 compared to WT or dBRCT; † p<.0001 compared to WT or dBRCT, p = .02 compared to dAMD. There is no significant difference between WT and dBRCT. (D) Frequencies of gene conversion events as a proportion of total mutations at the IgL locus (+/− SEM). n = total number of mutations analyzed for each cell line.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol (2010)
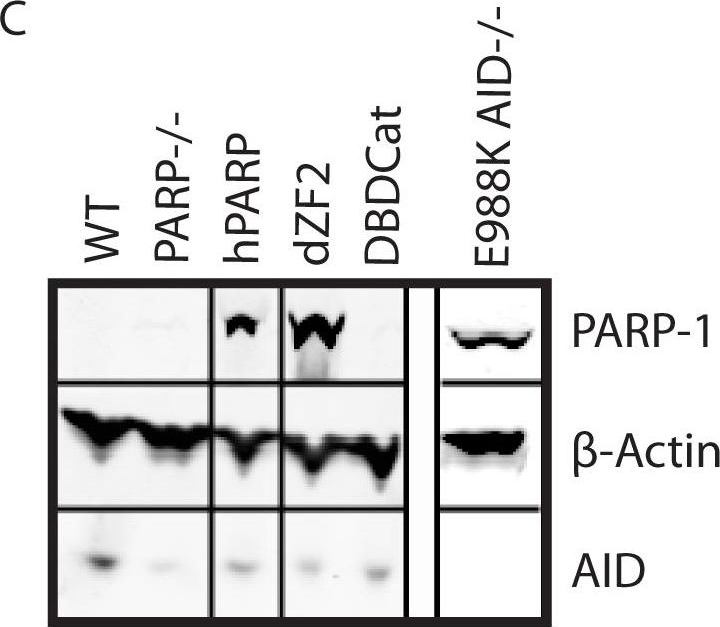
Functional effects of expression of human PARP-1 variants on survival in response to MMS-induced DNA damage.(A) Schematic of domains of human PARP-1 and variants. The functional domains of PARP-1 consist of a DNA binding domain (DBD), automodification domain (AMD), BRCT protein interaction domain (BRCT), and WGR/catalytic domain (WGR/Cat). The DBD contains 3 zinc finger domains, which are unusual in that they have specificity for DNA structure rather than sequence and recognize single strand breaks (SSBs) or double strand breaks (DSBs) [39],[40]. The AMD contains the lysine residues that act as poly-ADP-ribose (PAR) acceptors [35]. The WGR/catalytic domain catalyzes PAR formation when the DBD is bound to DNA, and PARylation of the AMD is thought to serve as a signal to recruit DNA repair enzymes such as XRCC1 as well as facilitates the release of PARP-1 from the site of DNA damage [41]. The BRCT protein interaction domain is of unknown function, as it has been shown to be dispensable for PARP-1’s DNA repair functions in previous analyses [16]. hPARP: full length human PARP-1; dZF2: C125Y and C128Y mutations to prevent folding of the second zinc finger domain; DBDCat: DNA binding domain fused to a non-functional portion of the catalytic domain. (B) MMS survival assay comparing survival of the PARP-1 variants to MMS-induced DNA damage. Survival is measured by the ability to proliferate after 1 h of exposure to MMS at the indicated concentration. The experiment was performed in triplicate; error bars represent SEM. *** PARP-1−/−, dZF2, and hPARP p<.0001 compared to WT; PARP-1−/− p<.0003 compared to hPARP; † p<.0001 compared to WT, p = .021 compared to PARP-1−/−; between PARP-1−/− and dZF2 there is no significant difference. (C) Western blot showing levels of variant PARP-1 and AID expression with β actin as a loading control.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol (2010)
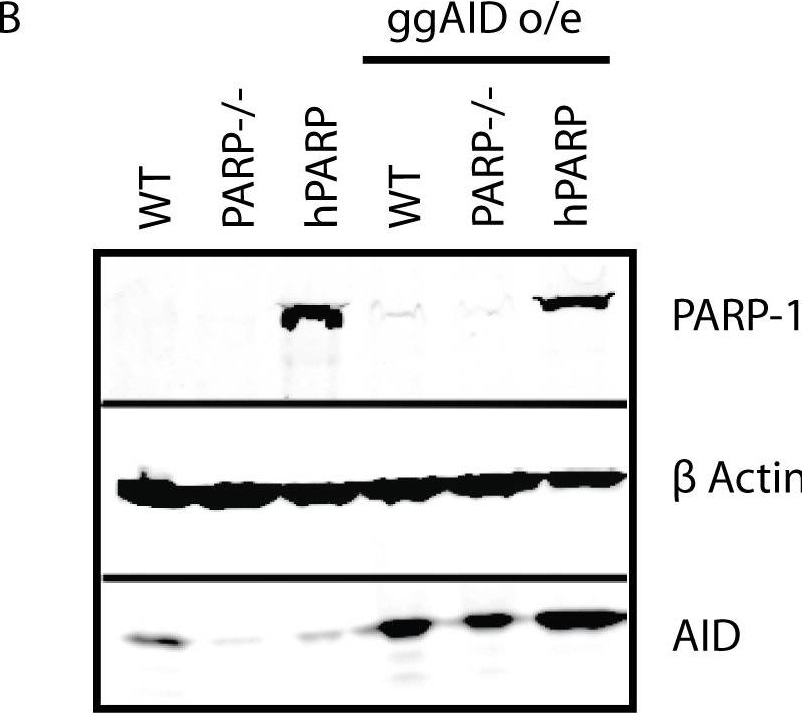
AID overexpression does not restore GCV to PARP-1−/− cells.(A) Gene conversion frequencies (+/− SEM) in cell lines overexpressing ggAID. n = total number of mutations analyzed for each cell line. The total number of sequences analyzed was 169 WT, 137 PARP-1−/−, and 106 hPARP. (B) Western blot showing increase in AID expression upon transduction with ggAID retrovirus. (C) IgL transcript levels (mean +/− SEM) are similar in cell lines which do and do not support GCV. * p<.05, ns = not significant compared to hPARP. (D) AID expression levels do not directly influence GCV frequencies. Blue bars are AID expression levels (mean +/− SEM) before (dark blue) and after (light blue) transduction with ggAID cDNA as measured by Western blot and quantified by LICOR Odessey infrared imaging, normalized to β actin. Brown bars are GCV frequencies (mean +/− SEM) before (dark brown) and after (light brown) transduction with ggAID cDNA as a percentage of total mutations observed for the indicated cell lines. *** p<.0001, * p<.05, ns = not significant.
Image collected and cropped by CiteAb under a CC-BY license from the following publication: The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol (2010)




Product Details
| Alternative Name |
Poly(ADP-ribose) polymerase-1 |
|---|---|
| Application |
ELISA, ICC, IHC (FS), IHC (PS), IP, WB |
| Application Notes |
Detects bands of ~116kDa (PARP-1) and ~85kDa (apoptosis-induced cleavage fragment) by Western blot. |
| Crossreactivity |
Does not cross-react with PARP-2. |
| Formulation |
Liquid. Neat serum containing 0.02% sodium azide. |
| Host |
Rabbit |
| Immunogen |
Recombinant human PARP-1 (poly(ADP-ribose) polymerase-1) (aa 1-1014). |
| Recommendation Dilutions/Conditions |
Immunocytochemistry (1:4,000)Immunoprecipitation (1:400)Western Blot (1:4,000)Suggested dilutions/conditions may not be available for all applications.Optimal conditions must be determined individually for each application. |
| Species Reactivity |
Bovine, Human, Monkey, Mouse |
| UniProt ID |
P09874 |
| Worry-free Guarantee |
This antibody is covered by our Worry-Free Guarantee. |
Handling & Storage
| Use/Stability |
Stable for at least one year when stored at +4°C. |
|---|---|
| Handling |
Avoid freeze/thaw cycles. |
| Long Term Storage |
+4°C |
| Shipping |
Blue Ice |
| Regulatory Status |
RUO – Research Use Only |
|---|
- Synergistic enhancement of PARP inhibition via small molecule UNI66-mediated suppression of BRD4-dependent transcription of RAD51 and CtIP: Amarsanaa, E., Wie, M., et al.; NAR Cancer 7, zcaf013 (2025), Abstract
- PARP1 associates with R-loops to promote their resolution and genome stability: Laspata, N., Kaur, P., et al.; Nucleic Acids Res. 51, 2215 (2023), Abstract
- Alkylation of nucleobases by 2-chloro-N,N-diethylethanamine hydrochloride (CDEAH) sensitizes PARP1-deficient tumors: Wie, M., Khim, K. W., et al.; NAR Cancer 5, zcad042 (2023), Abstract
- Cytidine deaminase deficiency in mice enhances genetic instability but limits the number of chemically induced colon tumors: R.O. Delic, et al.; Cancer Lett. 555, 216030 (2023), Abstract
- MDM2 binds and ubiquitinates PARP1 to enhance DNA replication fork progression: C. Giansanti, et al.; Cell Rep. 39, 110879 (2022), Abstract
- Epigenetic Landscape of HIV-1 Infection in Primary Human Macrophage: Lu, F., Zankharia, U., et al.; J. Virol. 96, e0016222 (2022), Abstract
- Poly(ADP) ribose polymerase promotes DNA polymerase theta-mediated end joining by activation of end resection: Ramsden, D., Feng, W., et al.; Research Square , (2022), Application(s): WB
- Low Sulfur Amino Acid, High Polyunsaturated Fatty Acid Diet Inhibits Breast Cancer Growth.: Turchi, R., Tortolici, F., et al.; Int. J. Mol. Sci. 24, (2022), Application(s): WB, Abstract
- Cytidine deaminase deficiency in tumor cells is associated with sensitivity to a naphthol derivative and a decrease in oncometabolite levels: H. Mameri, et al.; Cell. Mol. Life Sci. 79, 465 (2022), Application(s): WB, Abstract
- Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation.: Lieberman, P. M., Deng, Z., et al.; PLoS Pathog. 17, e1009231 (2021), Reactant(s): Human, Abstract
- Poly(ADP-ribosyl)ation temporally confines SUMO-dependent ataxin-3 recruitment to control DNA double-strand break repair.: Luijsterburg, M. S., Dantuma, N. P., et al.; J. Cell Sci. 134, (2021), Reactant(s): Human, Abstract
- Differential data on the responsiveness of multiple cell types to cell death induced by non-thermal atmospheric pressure plasma-activated solutions: K. Eto, et al.; Data Brief 36, 106995 (2021), Abstract — Full Text
- Topoisomerase Assays: Nitiss, J. L., Kiianitsa, K., et al.; Curr. Protoc. 1, e250 (2021), Abstract
- Treatment of human cells with 5-aza-dC induces formation of PARP1-DNA covalent adducts at genomic regions targeted by DNMT1.: Maizels, N., Kiianitsa, K., et al.; DNA Repair (Amst.) 96, 102977 (2020), Reactant(s): Human, Abstract
- Poly(ADP-ribose) Polymerase 1 (PARP1) restrains MyoD-dependent gene expression during muscle differentiation.: Matteini, F., Andresini, O., et al.; Sci. Rep. 10, 15086 (2020), Application(s): ChIP, Abstract
- A decrease in NAMPT activity impairs basal PARP-1 activity in cytidine deaminase deficient-cells, independently of NAD+: S. Silveira, et al.; Sci. Rep. 10, 13907 (2020), Application(s): WB / Reactant(s) Human, Abstract — Full Text
- Treatment of human cells with 5-aza-dC induces formation of PARP1-DNA covalent adducts at genomic regions targeted by DNMT1: Maizels, N., Kiianitsa, K., et al.; bioRxiv , (2019), Application(s): WB / Reactant(s): Human
- Epigenetic Regulation of RIP3 Suppresses Necroptosis and Increases Resistance to Chemotherapy in NonSmall Cell Lung Cancer: Q. Wang, et al.; Transl. Oncol. 13, 372 (2019), Abstract — Full Text
- Common Regulatory Pathways Mediate Activity of MicroRNAs Inducing Cardiomyocyte Proliferation: Torrini, C., Cubero, R. J., et al.; Cell Rep. 27, 2759 (2019), Abstract
- Cellular TRIM33 restrains HIV-1 infection by targeting viral integrase for proteasomal degradation: H. Ali, et al.; Nat. Commun. 10, 926 (2019), Abstract — Full Text
- Posttranscriptional Regulation of HIV-1 Gene Expression during Replication and Reactivation from Latency by Nuclear Matrix Protein MATR3: A. Sarracino, et al.; Mbio 9, e02158-18 (2018), Application(s): WB, Abstract — Full Text
- PARP1-Erk synergism in proliferating cells.: Visochek, L., Cohen-Armon, M., et al.; Oncotarget 9, 29140 (2018), Reactant(s): Mouse, Abstract
- PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR: Y. Ke, et al.; Nat. Commun. 8, 14632 (2017), Abstract — Full Text
- Cytidine deaminase deficiency impairs sister chromatid disjunction by decreasing PARP-1 activity: S. Gemble, et al.; Cell Cycle 16, 1128 (2017), Application(s): WB / Reactant(s) Human, Abstract — Full Text
- A PARP1-ERK2 synergism is required for the induction of LTP: L. Visochek, et al.; Sci. Rep. 6, 24950 (2016), Application(s): Western blot, Abstract — Full Text
- A balanced pyrimidine pool is required for optimal Chk1 activation to prevent ultrafine anaphase bridge formation: S. Gemble, et al.; J. Cell Sci. 129, 3167 (2016), Application(s): WB / Reactant(s) Human, Abstract
- NF-κB transcriptional activation by TNFα requires phospholipase C, extracellular signal-regulated kinase 2 and poly(ADP-ribose) polymerase-1: B. Vuong, et al.; J. Neuroninflammation 12, 229 (2015), Application(s): Western blot detecting PARP-1 expression, Abstract — Full Text
- Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB: D.L. Medina, et al.; Nat. Cell. Biol. 17, 288 (2015), Application(s): Western Blotting, Abstract
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA: S. Gemble, et al.; PLoS Genet. 11, 1005384 (2015), Application(s): WB / Reactant(s) Human, Abstract — Full Text
- A JNK-mediated autophagy pathway that triggers c-IAP degradation and necroptosis for anticancer chemotherapy: W. He, et al.; Oncogene 33, 3004 (2014), Reactant(s) Human, Abstract — Full Text
- Poly(ADP-Ribosyl)ation Is Required to Modulate Chromatin Changes at c-MYC Promoter during Emergence from Quiescence: C. Mostocotto, et al.; PLoS One 9, e102575 (2014), Application(s): ChIP, Abstract — Full Text
- Receptor-interacting Protein 1 Increases Chemoresistance by Maintaining Inhibitor of Apoptosis Protein Levels and Reducing Reactive Oxygen Species through a microRNA-146a-mediated Catalase Pathway: Q. Wang, et al.; J. Biol. Chem. 289, 5654 (2014), Reactant(s) Human, Abstract — Full Text
- Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition.: Mendoza-Maldonado, R., Aebersold, R., et al.; Nat. Struct. Mol. Biol. 20, 347 (2013), Reactant(s): Human, Abstract
- Poly(ADP-ribose) Polymerase 1 (PARP-1) Binds to 8-Oxoguanine-DNA Glycosylase (OGG1): N. Noren Hooten, et al.; J. Biol. Chem. 286, 44679 (2011), Application(s): IP, Abstract — Full Text
- RANKL up-regulates brain-type creatine kinase via poly(ADP-ribose) polymerase-1 during osteoclastogenesis: J. Chen, et al.; J. Biol. Chem. 285, 36315 (2010), Abstract — Full Text
- The BRCT domain of PARP-1 is required for immunoglobulin gene conversion: M.N. Paddock, et al.; PLoS Biol. 8, e1000428 (2010), Application(s): WB / Reactant(s) Chicken, Abstract — Full Text
- Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity: M.N. Rossi, et al.; J. Biol. Chem. 284, 31616 (2009), Application(s): IP, Abstract — Full Text
- Poly(adp-ribose) polymerase-1 regulates Tracp gene promoter activity during RANKL-induced osteoclastogenesis: G.E. Beranger, et al.; J. Bone Miner. Res. 23, 564 (2008), Abstract
- The prevention of spontaneous apoptosis of follicular lymphoma B cells by a follicular dendritic cell line: involvement of caspase-3, caspase-8 and c-FLIP: J.J. Goval, et al.; Haematologica 93, 1169 (2008), Application(s): WB using human follicular lymphoma lysate, Abstract — Full Text
- Differential binding of poly(ADP-Ribose) polymerase-1 and JunD/Fra2 accounts for RANKL-induced Tcirg1 gene expression during osteoclastogenesis: Beranger, G. E., Momier, D., et al.; J. Bone Miner. Res. 22, 975 (2007), Abstract
- RANKL treatment releases the negative regulation of the poly(ADP-ribose) polymerase-1 on Tcirg1 gene expression during osteoclastogenesis: Beranger, G. E., Momier, D., et al.; J. Bone Miner. Res. 21, 1757 (2006), Abstract
Datasheet, Manuals, SDS & CofA
Manuals And Inserts
Certificate of Analysis
Please enter the lot number as featured on the product label
SDS
Enzo Life Science provides GHS Compliant SDS
If your language is not available please fill out the SDS request form
 AMPIVIEW® RNA probes
AMPIVIEW® RNA probes Enabling Your Projects
Enabling Your Projects  GMP Services
GMP Services Bulk Solutions
Bulk Solutions Research Travel Grant
Research Travel Grant Have You Published Using an Enzo Product?
Have You Published Using an Enzo Product?
